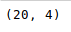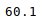Excel sheets are very instinctive and user-friendly, which makes them ideal for manipulating large datasets even for less technical folks. If you are looking for places to learn to manipulate and automate stuff in excel files using Python, look no more. You are at the right place.
Python Pandas With Excel Sheet
In this article, you will learn how to use Pandas to work with Excel spreadsheets. At the end of the article, you will have the knowledge of:
- Necessary modules are needed for this and how to set them up in your system.
- Reading data from excel files into pandas using Python.
- Exploring the data from excel files in Pandas.
- Using functions to manipulate and reshape the data in Pandas.
Installation
To install Pandas in Anaconda, we can use the following command in Anaconda Terminal:
conda install pandas
To install Pandas in regular Python (Non-Anaconda), we can use the following command in the command prompt:
pip install pandas
Getting Started
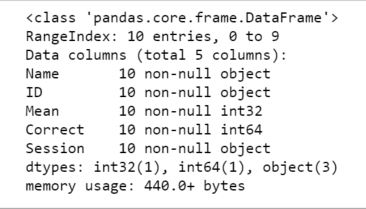
First of all, we need to import the Pandas module which can be done by running the command: Pandas
Python3
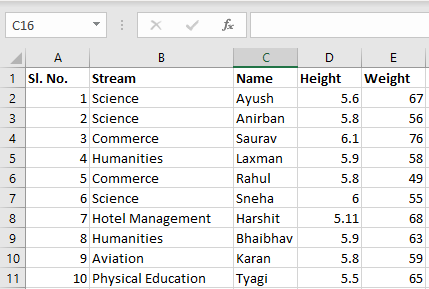
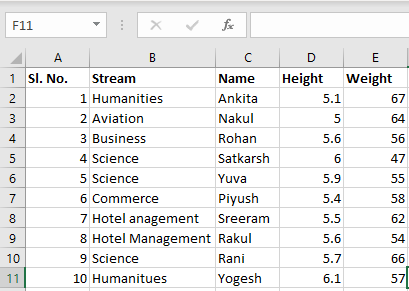
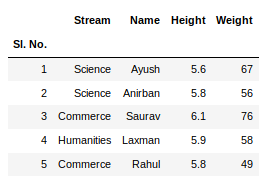
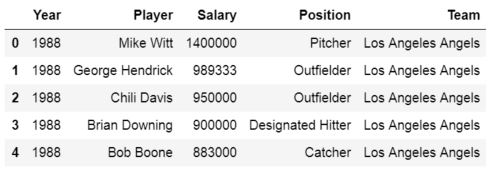
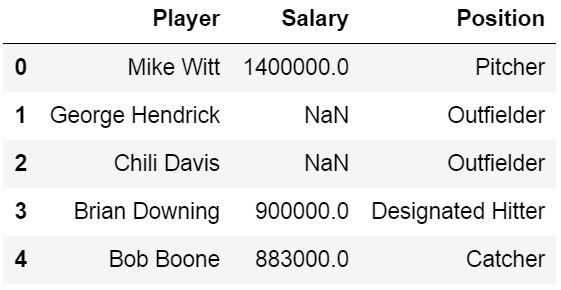
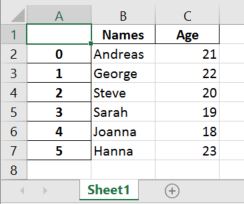
Input File: Let’s suppose the excel file looks like this
Sheet 1:
Sheet 2:
Now we can import the excel file using the read_excel function in Pandas. The second statement reads the data from excel and stores it into a pandas Data Frame which is represented by the variable newData. If there are multiple sheets in the excel workbook, the command will import data of the first sheet. To make a data frame with all the sheets in the workbook, the easiest method is to create different data frames separately and then concatenate them. The read_excel method takes argument sheet_name and index_col where we can specify the sheet of which the data frame should be made of and index_col specifies the title column, as is shown below:
Python3
file =('path_of_excel_file')
newData = pds.read_excel(file)
newData
Output:
Example:
The third statement concatenates both sheets. Now to check the whole data frame, we can simply run the following command:
Python3
sheet1 = pds.read_excel(file,
sheet_name = 0,
index_col = 0)
sheet2 = pds.read_excel(file,
sheet_name = 1,
index_col = 0)
newData = pds.concat([sheet1, sheet2])
newData
Output:
To view 5 columns from the top and from the bottom of the data frame, we can run the command. This head() and tail() method also take arguments as numbers for the number of columns to show.
Python3
newData.head()
newData.tail()
Output:
The shape() method can be used to view the number of rows and columns in the data frame as follows:
Python3
Output:
If any column contains numerical data, we can sort that column using the sort_values() method in pandas as follows:
Python3
sorted_column = newData.sort_values(['Height'], ascending = False)
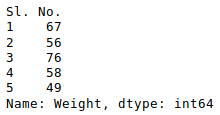
Now, let’s suppose we want the top 5 values of the sorted column, we can use the head() method here:
Python3
sorted_column['Height'].head(5)
Output:
We can do that with any numerical column of the data frame as shown below:
Python3
Output:
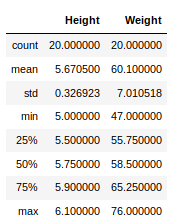
Now, suppose our data is mostly numerical. We can get the statistical information like mean, max, min, etc. about the data frame using the describe() method as shown below:
Python3
Output:
This can also be done separately for all the numerical columns using the following command:
Python3
Output:
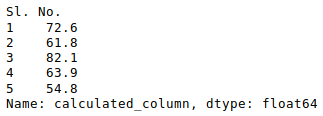
Other statistical information can also be calculated using the respective methods. Like in excel, formulas can also be applied and calculated columns can be created as follows:
Python3
newData['calculated_column'] =
newData[“Height”] + newData[“Weight”]
newData['calculated_column'].head()
Output:
After operating on the data in the data frame, we can export the data back to an excel file using the method to_excel. For this we need to specify an output excel file where the transformed data is to be written, as shown below:
Python3
newData.to_excel('Output File.xlsx')
Output:
The read_excel() method can read Excel 2003 (.xls) and
Excel 2007+ (.xlsx) files using the xlrd Python
module. The to_excel() instance method is used for
saving a DataFrame to Excel. Generally the semantics are
similar to working with csv data. See the cookbook for some
advanced strategies
10.5.1 Reading Excel Files
In the most basic use-case, read_excel takes a path to an Excel
file, and the sheetname indicating which sheet to parse.
# Returns a DataFrame read_excel('path_to_file.xls', sheetname='Sheet1')
10.5.1.1 ExcelFile class
To facilitate working with multiple sheets from the same file, the ExcelFile
class can be used to wrap the file and can be be passed into read_excel
There will be a performance benefit for reading multiple sheets as the file is
read into memory only once.
xlsx = pd.ExcelFile('path_to_file.xls) df = pd.read_excel(xlsx, 'Sheet1')
The ExcelFile class can also be used as a context manager.
with pd.ExcelFile('path_to_file.xls') as xls: df1 = pd.read_excel(xls, 'Sheet1') df2 = pd.read_excel(xls, 'Sheet2')
The sheet_names property will generate
a list of the sheet names in the file.
The primary use-case for an ExcelFile is parsing multiple sheets with
different parameters
data = {} # For when Sheet1's format differs from Sheet2 with pd.ExcelFile('path_to_file.xls') as xls: data['Sheet1'] = pd.read_excel(xls, 'Sheet1', index_col=None, na_values=['NA']) data['Sheet2'] = pd.read_excel(xls, 'Sheet2', index_col=1)
Note that if the same parsing parameters are used for all sheets, a list
of sheet names can simply be passed to read_excel with no loss in performance.
# using the ExcelFile class data = {} with pd.ExcelFile('path_to_file.xls') as xls: data['Sheet1'] = read_excel(xls, 'Sheet1', index_col=None, na_values=['NA']) data['Sheet2'] = read_excel(xls, 'Sheet2', index_col=None, na_values=['NA']) # equivalent using the read_excel function data = read_excel('path_to_file.xls', ['Sheet1', 'Sheet2'], index_col=None, na_values=['NA'])
New in version 0.12.
ExcelFile has been moved to the top level namespace.
New in version 0.17.
read_excel can take an ExcelFile object as input
10.5.1.2 Specifying Sheets
Note
The second argument is sheetname, not to be confused with ExcelFile.sheet_names
Note
An ExcelFile’s attribute sheet_names provides access to a list of sheets.
- The arguments
sheetnameallows specifying the sheet or sheets to read. - The default value for
sheetnameis 0, indicating to read the first sheet - Pass a string to refer to the name of a particular sheet in the workbook.
- Pass an integer to refer to the index of a sheet. Indices follow Python
convention, beginning at 0. - Pass a list of either strings or integers, to return a dictionary of specified sheets.
- Pass a
Noneto return a dictionary of all available sheets.
# Returns a DataFrame read_excel('path_to_file.xls', 'Sheet1', index_col=None, na_values=['NA'])
Using the sheet index:
# Returns a DataFrame read_excel('path_to_file.xls', 0, index_col=None, na_values=['NA'])
Using all default values:
# Returns a DataFrame read_excel('path_to_file.xls')
Using None to get all sheets:
# Returns a dictionary of DataFrames read_excel('path_to_file.xls',sheetname=None)
Using a list to get multiple sheets:
# Returns the 1st and 4th sheet, as a dictionary of DataFrames. read_excel('path_to_file.xls',sheetname=['Sheet1',3])
New in version 0.16.
read_excel can read more than one sheet, by setting sheetname to either
a list of sheet names, a list of sheet positions, or None to read all sheets.
New in version 0.13.
Sheets can be specified by sheet index or sheet name, using an integer or string,
respectively.
10.5.1.3 Reading a MultiIndex
New in version 0.17.
read_excel can read a MultiIndex index, by passing a list of columns to index_col
and a MultiIndex column by passing a list of rows to header. If either the index
or columns have serialized level names those will be read in as well by specifying
the rows/columns that make up the levels.
For example, to read in a MultiIndex index without names:
In [1]: df = pd.DataFrame({'a':[1,2,3,4], 'b':[5,6,7,8]}, ...: index=pd.MultiIndex.from_product([['a','b'],['c','d']])) ...: In [2]: df.to_excel('path_to_file.xlsx') In [3]: df = pd.read_excel('path_to_file.xlsx', index_col=[0,1]) In [4]: df Out[4]: a b a c 1 5 d 2 6 b c 3 7 d 4 8
If the index has level names, they will parsed as well, using the same
parameters.
In [5]: df.index = df.index.set_names(['lvl1', 'lvl2']) In [6]: df.to_excel('path_to_file.xlsx') In [7]: df = pd.read_excel('path_to_file.xlsx', index_col=[0,1]) In [8]: df Out[8]: a b lvl1 lvl2 a c 1 5 d 2 6 b c 3 7 d 4 8
If the source file has both MultiIndex index and columns, lists specifying each
should be passed to index_col and header
In [9]: df.columns = pd.MultiIndex.from_product([['a'],['b', 'd']], names=['c1', 'c2']) In [10]: df.to_excel('path_to_file.xlsx') In [11]: df = pd.read_excel('path_to_file.xlsx', ....: index_col=[0,1], header=[0,1]) ....: In [12]: df Out[12]: c1 a c2 b d lvl1 lvl2 a c 1 5 d 2 6 b c 3 7 d 4 8
Warning
Excel files saved in version 0.16.2 or prior that had index names will still able to be read in,
but the has_index_names argument must specified to True.
10.5.1.4 Parsing Specific Columns
It is often the case that users will insert columns to do temporary computations
in Excel and you may not want to read in those columns. read_excel takes
a parse_cols keyword to allow you to specify a subset of columns to parse.
If parse_cols is an integer, then it is assumed to indicate the last column
to be parsed.
read_excel('path_to_file.xls', 'Sheet1', parse_cols=2)
If parse_cols is a list of integers, then it is assumed to be the file column
indices to be parsed.
read_excel('path_to_file.xls', 'Sheet1', parse_cols=[0, 2, 3])
10.5.1.5 Cell Converters
It is possible to transform the contents of Excel cells via the converters
option. For instance, to convert a column to boolean:
read_excel('path_to_file.xls', 'Sheet1', converters={'MyBools': bool})
This options handles missing values and treats exceptions in the converters
as missing data. Transformations are applied cell by cell rather than to the
column as a whole, so the array dtype is not guaranteed. For instance, a
column of integers with missing values cannot be transformed to an array
with integer dtype, because NaN is strictly a float. You can manually mask
missing data to recover integer dtype:
cfun = lambda x: int(x) if x else -1 read_excel('path_to_file.xls', 'Sheet1', converters={'MyInts': cfun})
10.5.2 Writing Excel Files
10.5.2.1 Writing Excel Files to Disk
To write a DataFrame object to a sheet of an Excel file, you can use the
to_excel instance method. The arguments are largely the same as to_csv
described above, the first argument being the name of the excel file, and the
optional second argument the name of the sheet to which the DataFrame should be
written. For example:
df.to_excel('path_to_file.xlsx', sheet_name='Sheet1')
Files with a .xls extension will be written using xlwt and those with a
.xlsx extension will be written using xlsxwriter (if available) or
openpyxl.
The DataFrame will be written in a way that tries to mimic the REPL output. One
difference from 0.12.0 is that the index_label will be placed in the second
row instead of the first. You can get the previous behaviour by setting the
merge_cells option in to_excel() to False:
df.to_excel('path_to_file.xlsx', index_label='label', merge_cells=False)
The Panel class also has a to_excel instance method,
which writes each DataFrame in the Panel to a separate sheet.
In order to write separate DataFrames to separate sheets in a single Excel file,
one can pass an ExcelWriter.
with ExcelWriter('path_to_file.xlsx') as writer: df1.to_excel(writer, sheet_name='Sheet1') df2.to_excel(writer, sheet_name='Sheet2')
Note
Wringing a little more performance out of read_excel
Internally, Excel stores all numeric data as floats. Because this can
produce unexpected behavior when reading in data, pandas defaults to trying
to convert integers to floats if it doesn’t lose information (1.0 -->). You can pass
1convert_float=False to disable this behavior, which
may give a slight performance improvement.
10.5.2.2 Writing Excel Files to Memory
New in version 0.17.
Pandas supports writing Excel files to buffer-like objects such as StringIO or
BytesIO using ExcelWriter.
New in version 0.17.
Added support for Openpyxl >= 2.2
# Safe import for either Python 2.x or 3.x try: from io import BytesIO except ImportError: from cStringIO import StringIO as BytesIO bio = BytesIO() # By setting the 'engine' in the ExcelWriter constructor. writer = ExcelWriter(bio, engine='xlsxwriter') df.to_excel(writer, sheet_name='Sheet1') # Save the workbook writer.save() # Seek to the beginning and read to copy the workbook to a variable in memory bio.seek(0) workbook = bio.read()
Note
engine is optional but recommended. Setting the engine determines
the version of workbook produced. Setting engine='xlrd' will produce an
Excel 2003-format workbook (xls). Using either 'openpyxl' or
'xlsxwriter' will produce an Excel 2007-format workbook (xlsx). If
omitted, an Excel 2007-formatted workbook is produced.
10.5.3 Excel writer engines
New in version 0.13.
pandas chooses an Excel writer via two methods:
- the
enginekeyword argument - the filename extension (via the default specified in config options)
By default, pandas uses the XlsxWriter for .xlsx and openpyxl
for .xlsm files and xlwt for .xls files. If you have multiple
engines installed, you can set the default engine through setting the
config options io.excel.xlsx.writer and
io.excel.xls.writer. pandas will fall back on openpyxl for .xlsx
files if Xlsxwriter is not available.
To specify which writer you want to use, you can pass an engine keyword
argument to to_excel and to ExcelWriter. The built-in engines are:
openpyxl: This includes stable support for Openpyxl from 1.6.1. However,
it is advised to use version 2.2 and higher, especially when working with
styles.xlsxwriterxlwt
# By setting the 'engine' in the DataFrame and Panel 'to_excel()' methods. df.to_excel('path_to_file.xlsx', sheet_name='Sheet1', engine='xlsxwriter') # By setting the 'engine' in the ExcelWriter constructor. writer = ExcelWriter('path_to_file.xlsx', engine='xlsxwriter') # Or via pandas configuration. from pandas import options options.io.excel.xlsx.writer = 'xlsxwriter' df.to_excel('path_to_file.xlsx', sheet_name='Sheet1')
Why learn to work with Excel with Python? Excel is one of the most popular and widely-used data tools; it’s hard to find an organization that doesn’t work with it in some way. From analysts, to sales VPs, to CEOs, various professionals use Excel for both quick stats and serious data crunching.
With Excel being so pervasive, data professionals must be familiar with it. Working with data in Python or R offers serious advantages over Excel’s UI, so finding a way to work with Excel using code is critical. Thankfully, there’s a great tool already out there for using Excel with Python called pandas.
Pandas has excellent methods for reading all kinds of data from Excel files. You can also export your results from pandas back to Excel, if that’s preferred by your intended audience. Pandas is great for other routine data analysis tasks, such as:
- quick Exploratory Data Analysis (EDA)
- drawing attractive plots
- feeding data into machine learning tools like scikit-learn
- building machine learning models on your data
- taking cleaned and processed data to any number of data tools
Pandas is better at automating data processing tasks than Excel, including processing Excel files.
In this tutorial, we are going to show you how to work with Excel files in pandas. We will cover the following concepts.
- setting up your computer with the necessary software
- reading in data from Excel files into pandas
- data exploration in pandas
- visualizing data in pandas using the matplotlib visualization library
- manipulating and reshaping data in pandas
- moving data from pandas into Excel
Note that this tutorial does not provide a deep dive into pandas. To explore pandas more, check out our course.
System Prerequisites
We will use Python 3 and Jupyter Notebook to demonstrate the code in this tutorial.In addition to Python and Jupyter Notebook, you will need the following Python modules:
- matplotlib — data visualization
- NumPy — numerical data functionality
- OpenPyXL — read/write Excel 2010 xlsx/xlsm files
- pandas — data import, clean-up, exploration, and analysis
- xlrd — read Excel data
- xlwt — write to Excel
- XlsxWriter — write to Excel (xlsx) files
There are multiple ways to get set up with all the modules. We cover three of the most common scenarios below.
- If you have Python installed via Anaconda package manager, you can install the required modules using the command
conda install. For example, to install pandas, you would execute the command —conda install pandas. - If you already have a regular, non-Anaconda Python installed on the computer, you can install the required modules using
pip. Open your command line program and execute commandpip install <module name>to install a module. You should replace<module name>with the actual name of the module you are trying to install. For example, to install pandas, you would execute command —pip install pandas. - If you don’t have Python already installed, you should get it through the Anaconda package manager. Anaconda provides installers for Windows, Mac, and Linux Computers. If you choose the full installer, you will get all the modules you need, along with Python and pandas within a single package. This is the easiest and fastest way to get started.
The Data Set
In this tutorial, we will use a multi-sheet Excel file we created from Kaggle’s IMDB Scores data. You can download the file here.
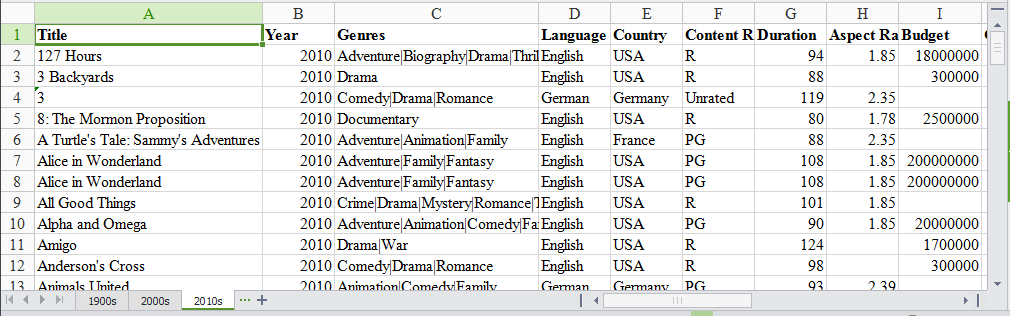
Our Excel file has three sheets: ‘1900s,’ ‘2000s,’ and ‘2010s.’ Each sheet has data for movies from those years.
We will use this data set to find the ratings distribution for the movies, visualize movies with highest ratings and net earnings and calculate statistical information about the movies. We will be analyzing and exploring this data using Python and pandas, thus demonstrating pandas capabilities for working with Excel data in Python.
Read data from the Excel file
We need to first import the data from the Excel file into pandas. To do that, we start by importing the pandas module.
import pandas as pdWe then use the pandas’ read_excel method to read in data from the Excel file. The easiest way to call this method is to pass the file name. If no sheet name is specified then it will read the first sheet in the index (as shown below).
excel_file = 'movies.xls'
movies = pd.read_excel(excel_file)Here, the read_excel method read the data from the Excel file into a pandas DataFrame object. Pandas defaults to storing data in DataFrames. We then stored this DataFrame into a variable called movies.
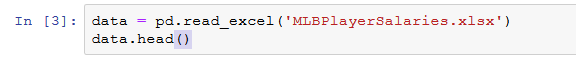
Pandas has a built-in DataFrame.head() method that we can use to easily display the first few rows of our DataFrame. If no argument is passed, it will display first five rows. If a number is passed, it will display the equal number of rows from the top.
movies.head()| Title | Year | Genres | Language | Country | Content Rating | Duration | Aspect Ratio | Budget | Gross Earnings | … | Facebook Likes — Actor 1 | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Intolerance: Love’s Struggle Throughout the Ages | 1916 | Drama|History|War | NaN | USA | Not Rated | 123 | 1.33 | 385907.0 | NaN | … | 436 | 22 | 9.0 | 481 | 691 | 1 | 10718 | 88 | 69.0 | 8.0 |
| 1 | Over the Hill to the Poorhouse | 1920 | Crime|Drama | NaN | USA | NaN | 110 | 1.33 | 100000.0 | 3000000.0 | … | 2 | 2 | 0.0 | 4 | 0 | 1 | 5 | 1 | 1.0 | 4.8 |
| 2 | The Big Parade | 1925 | Drama|Romance|War | NaN | USA | Not Rated | 151 | 1.33 | 245000.0 | NaN | … | 81 | 12 | 6.0 | 108 | 226 | 0 | 4849 | 45 | 48.0 | 8.3 |
| 3 | Metropolis | 1927 | Drama|Sci-Fi | German | Germany | Not Rated | 145 | 1.33 | 6000000.0 | 26435.0 | … | 136 | 23 | 18.0 | 203 | 12000 | 1 | 111841 | 413 | 260.0 | 8.3 |
| 4 | Pandora’s Box | 1929 | Crime|Drama|Romance | German | Germany | Not Rated | 110 | 1.33 | NaN | 9950.0 | … | 426 | 20 | 3.0 | 455 | 926 | 1 | 7431 | 84 | 71.0 | 8.0 |
5 rows × 25 columns
Excel files quite often have multiple sheets and the ability to read a specific sheet or all of them is very important. To make this easy, the pandas read_excel method takes an argument called sheetname that tells pandas which sheet to read in the data from. For this, you can either use the sheet name or the sheet number. Sheet numbers start with zero. If the sheetname argument is not given, it defaults to zero and pandas will import the first sheet.
By default, pandas will automatically assign a numeric index or row label starting with zero. You may want to leave the default index as such if your data doesn’t have a column with unique values that can serve as a better index. In case there is a column that you feel would serve as a better index, you can override the default behavior by setting index_col property to a column. It takes a numeric value for setting a single column as index or a list of numeric values for creating a multi-index.
In the below code, we are choosing the first column, ‘Title’, as index (index=0) by passing zero to the index_col argument.
movies_sheet1 = pd.read_excel(excel_file, sheetname=0, index_col=0)
movies_sheet1.head()| Year | Genres | Language | Country | Content Rating | Duration | Aspect Ratio | Budget | Gross Earnings | Director | … | Facebook Likes — Actor 1 | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | |||||||||||||||||||||
| Intolerance: Love’s Struggle Throughout the Ages | 1916 | Drama|History|War | NaN | USA | Not Rated | 123 | 1.33 | 385907.0 | NaN | D.W. Griffith | … | 436 | 22 | 9.0 | 481 | 691 | 1 | 10718 | 88 | 69.0 | 8.0 |
| Over the Hill to the Poorhouse | 1920 | Crime|Drama | NaN | USA | NaN | 110 | 1.33 | 100000.0 | 3000000.0 | Harry F. Millarde | … | 2 | 2 | 0.0 | 4 | 0 | 1 | 5 | 1 | 1.0 | 4.8 |
| The Big Parade | 1925 | Drama|Romance|War | NaN | USA | Not Rated | 151 | 1.33 | 245000.0 | NaN | King Vidor | … | 81 | 12 | 6.0 | 108 | 226 | 0 | 4849 | 45 | 48.0 | 8.3 |
| Metropolis | 1927 | Drama|Sci-Fi | German | Germany | Not Rated | 145 | 1.33 | 6000000.0 | 26435.0 | Fritz Lang | … | 136 | 23 | 18.0 | 203 | 12000 | 1 | 111841 | 413 | 260.0 | 8.3 |
| Pandora’s Box | 1929 | Crime|Drama|Romance | German | Germany | Not Rated | 110 | 1.33 | NaN | 9950.0 | Georg Wilhelm Pabst | … | 426 | 20 | 3.0 | 455 | 926 | 1 | 7431 | 84 | 71.0 | 8.0 |
5 rows × 24 columns
As you noticed above, our Excel data file has three sheets. We already read the first sheet in a DataFrame above. Now, using the same syntax, we will read in rest of the two sheets too.
movies_sheet2 = pd.read_excel(excel_file, sheetname=1, index_col=0)
movies_sheet2.head()| Year | Genres | Language | Country | Content Rating | Duration | Aspect Ratio | Budget | Gross Earnings | Director | … | Facebook Likes — Actor 1 | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | |||||||||||||||||||||
| 102 Dalmatians | 2000 | Adventure|Comedy|Family | English | USA | G | 100.0 | 1.85 | 85000000.0 | 66941559.0 | Kevin Lima | … | 2000.0 | 795.0 | 439.0 | 4182 | 372 | 1 | 26413 | 77.0 | 84.0 | 4.8 |
| 28 Days | 2000 | Comedy|Drama | English | USA | PG-13 | 103.0 | 1.37 | 43000000.0 | 37035515.0 | Betty Thomas | … | 12000.0 | 10000.0 | 664.0 | 23864 | 0 | 1 | 34597 | 194.0 | 116.0 | 6.0 |
| 3 Strikes | 2000 | Comedy | English | USA | R | 82.0 | 1.85 | 6000000.0 | 9821335.0 | DJ Pooh | … | 939.0 | 706.0 | 585.0 | 3354 | 118 | 1 | 1415 | 10.0 | 22.0 | 4.0 |
| Aberdeen | 2000 | Drama | English | UK | NaN | 106.0 | 1.85 | 6500000.0 | 64148.0 | Hans Petter Moland | … | 844.0 | 2.0 | 0.0 | 846 | 260 | 0 | 2601 | 35.0 | 28.0 | 7.3 |
| All the Pretty Horses | 2000 | Drama|Romance|Western | English | USA | PG-13 | 220.0 | 2.35 | 57000000.0 | 15527125.0 | Billy Bob Thornton | … | 13000.0 | 861.0 | 820.0 | 15006 | 652 | 2 | 11388 | 183.0 | 85.0 | 5.8 |
5 rows × 24 columns
movies_sheet3 = pd.read_excel(excel_file, sheetname=2, index_col=0)
movies_sheet3.head()| Year | Genres | Language | Country | Content Rating | Duration | Aspect Ratio | Budget | Gross Earnings | Director | … | Facebook Likes — Actor 1 | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | |||||||||||||||||||||
| 127 Hours | 2010.0 | Adventure|Biography|Drama|Thriller | English | USA | R | 94.0 | 1.85 | 18000000.0 | 18329466.0 | Danny Boyle | … | 11000.0 | 642.0 | 223.0 | 11984 | 63000 | 0.0 | 279179 | 440.0 | 450.0 | 7.6 |
| 3 Backyards | 2010.0 | Drama | English | USA | R | 88.0 | NaN | 300000.0 | NaN | Eric Mendelsohn | … | 795.0 | 659.0 | 301.0 | 1884 | 92 | 0.0 | 554 | 23.0 | 20.0 | 5.2 |
| 3 | 2010.0 | Comedy|Drama|Romance | German | Germany | Unrated | 119.0 | 2.35 | NaN | 59774.0 | Tom Tykwer | … | 24.0 | 20.0 | 9.0 | 69 | 2000 | 0.0 | 4212 | 18.0 | 76.0 | 6.8 |
| 8: The Mormon Proposition | 2010.0 | Documentary | English | USA | R | 80.0 | 1.78 | 2500000.0 | 99851.0 | Reed Cowan | … | 191.0 | 12.0 | 5.0 | 210 | 0 | 0.0 | 1138 | 30.0 | 28.0 | 7.1 |
| A Turtle’s Tale: Sammy’s Adventures | 2010.0 | Adventure|Animation|Family | English | France | PG | 88.0 | 2.35 | NaN | NaN | Ben Stassen | … | 783.0 | 749.0 | 602.0 | 3874 | 0 | 2.0 | 5385 | 22.0 | 56.0 | 6.1 |
5 rows × 24 columns
Since all the three sheets have similar data but for different recordsmovies, we will create a single DataFrame from all the three DataFrames we created above. We will use the pandas concat method for this and pass in the names of the three DataFrames we just created and assign the results to a new DataFrame object, movies. By keeping the DataFrame name same as before, we are over-writing the previously created DataFrame.
movies = pd.concat([movies_sheet1, movies_sheet2, movies_sheet3])We can check if this concatenation by checking the number of rows in the combined DataFrame by calling the method shape on it that will give us the number of rows and columns.
movies.shape(5042, 24)Using the ExcelFile class to read multiple sheets
We can also use the ExcelFile class to work with multiple sheets from the same Excel file. We first wrap the Excel file using ExcelFile and then pass it to read_excel method.
xlsx = pd.ExcelFile(excel_file)
movies_sheets = []
for sheet in xlsx.sheet_names:
movies_sheets.append(xlsx.parse(sheet))
movies = pd.concat(movies_sheets)If you are reading an Excel file with a lot of sheets and are creating a lot of DataFrames, ExcelFile is more convenient and efficient in comparison to read_excel. With ExcelFile, you only need to pass the Excel file once, and then you can use it to get the DataFrames. When using read_excel, you pass the Excel file every time and hence the file is loaded again for every sheet. This can be a huge performance drag if the Excel file has many sheets with a large number of rows.
Exploring the data
Now that we have read in the movies data set from our Excel file, we can start exploring it using pandas. A pandas DataFrame stores the data in a tabular format, just like the way Excel displays the data in a sheet. Pandas has a lot of built-in methods to explore the DataFrame we created from the Excel file we just read in.
We already introduced the method head in the previous section that displays few rows from the top from the DataFrame. Let’s look at few more methods that come in handy while exploring the data set.
We can use the shape method to find out the number of rows and columns for the DataFrame.
movies.shape(5042, 25)This tells us our Excel file has 5042 records and 25 columns or observations. This can be useful in reporting the number of records and columns and comparing that with the source data set.
We can use the tail method to view the bottom rows. If no parameter is passed, only the bottom five rows are returned.
movies.tail()| Title | Year | Genres | Language | Country | Content Rating | Duration | Aspect Ratio | Budget | Gross Earnings | … | Facebook Likes — Actor 1 | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1599 | War & Peace | NaN | Drama|History|Romance|War | English | UK | TV-14 | NaN | 16.00 | NaN | NaN | … | 1000.0 | 888.0 | 502.0 | 4528 | 11000 | 1.0 | 9277 | 44.0 | 10.0 | 8.2 |
| 1600 | Wings | NaN | Comedy|Drama | English | USA | NaN | 30.0 | 1.33 | NaN | NaN | … | 685.0 | 511.0 | 424.0 | 1884 | 1000 | 5.0 | 7646 | 56.0 | 19.0 | 7.3 |
| 1601 | Wolf Creek | NaN | Drama|Horror|Thriller | English | Australia | NaN | NaN | 2.00 | NaN | NaN | … | 511.0 | 457.0 | 206.0 | 1617 | 954 | 0.0 | 726 | 6.0 | 2.0 | 7.1 |
| 1602 | Wuthering Heights | NaN | Drama|Romance | English | UK | NaN | 142.0 | NaN | NaN | NaN | … | 27000.0 | 698.0 | 427.0 | 29196 | 0 | 2.0 | 6053 | 33.0 | 9.0 | 7.7 |
| 1603 | Yu-Gi-Oh! Duel Monsters | NaN | Action|Adventure|Animation|Family|Fantasy | Japanese | Japan | NaN | 24.0 | NaN | NaN | NaN | … | 0.0 | NaN | NaN | 0 | 124 | 0.0 | 12417 | 51.0 | 6.0 | 7.0 |
5 rows × 25 columns
In Excel, you’re able to sort a sheet based on the values in one or more columns. In pandas, you can do the same thing with the sort_values method. For example, let’s sort our movies DataFrame based on the Gross Earnings column.
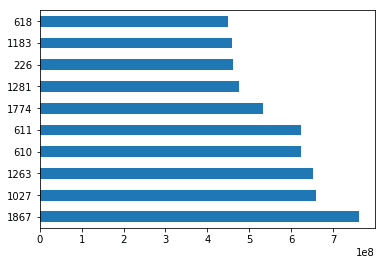
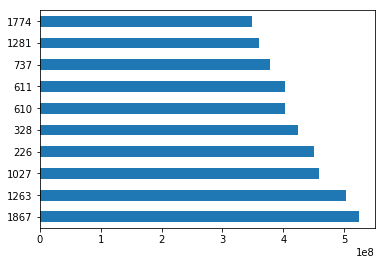
sorted_by_gross = movies.sort_values(['Gross Earnings'], ascending=False)Since we have the data sorted by values in a column, we can do few interesting things with it. For example, we can display the top 10 movies by Gross Earnings.
sorted_by_gross["Gross Earnings"].head(10)1867 760505847.0
1027 658672302.0
1263 652177271.0
610 623279547.0
611 623279547.0
1774 533316061.0
1281 474544677.0
226 460935665.0
1183 458991599.0
618 448130642.0
Name: Gross Earnings, dtype: float64We can also create a plot for the top 10 movies by Gross Earnings. Pandas makes it easy to visualize your data with plots and charts through matplotlib, a popular data visualization library. With a couple lines of code, you can start plotting. Moreover, matplotlib plots work well inside Jupyter Notebooks since you can displace the plots right under the code.
First, we import the matplotlib module and set matplotlib to display the plots right in the Jupyter Notebook.
import matplotlib.pyplot as plt%matplotlib inlineWe will draw a bar plot where each bar will represent one of the top 10 movies. We can do this by calling the plot method and setting the argument kind to barh. This tells matplotlib to draw a horizontal bar plot.
sorted_by_gross['Gross Earnings'].head(10).plot(kind="barh")
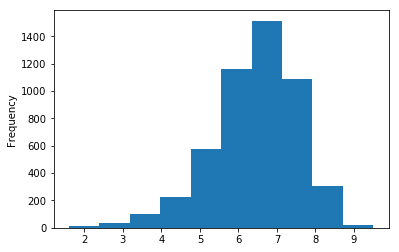
plt.show()Let’s create a histogram of IMDB Scores to check the distribution of IMDB Scores across all movies. Histograms are a good way to visualize the distribution of a data set. We use the plot method on the IMDB Scores series from our movies DataFrame and pass it the argument.
movies['IMDB Score'].plot(kind="hist")
plt.show()This data visualization suggests that most of the IMDB Scores fall between six and eight.
Getting statistical information about the data
Pandas has some very handy methods to look at the statistical data about our data set. For example, we can use the describe method to get a statistical summary of the data set.
movies.describe()| Year | Duration | Aspect Ratio | Budget | Gross Earnings | Facebook Likes — Director | Facebook Likes — Actor 1 | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 4935.000000 | 5028.000000 | 4714.000000 | 4.551000e+03 | 4.159000e+03 | 4938.000000 | 5035.000000 | 5029.000000 | 5020.000000 | 5042.000000 | 5042.000000 | 5029.000000 | 5.042000e+03 | 5022.000000 | 4993.000000 | 5042.000000 |
| mean | 2002.470517 | 107.201074 | 2.220403 | 3.975262e+07 | 4.846841e+07 | 686.621709 | 6561.323932 | 1652.080533 | 645.009761 | 9700.959143 | 7527.457160 | 1.371446 | 8.368475e+04 | 272.770808 | 140.194272 | 6.442007 |
| std | 12.474599 | 25.197441 | 1.385113 | 2.061149e+08 | 6.845299e+07 | 2813.602405 | 15021.977635 | 4042.774685 | 1665.041728 | 18165.101925 | 19322.070537 | 2.013683 | 1.384940e+05 | 377.982886 | 121.601675 | 1.125189 |
| min | 1916.000000 | 7.000000 | 1.180000 | 2.180000e+02 | 1.620000e+02 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 5.000000e+00 | 1.000000 | 1.000000 | 1.600000 |
| 25% | 1999.000000 | 93.000000 | 1.850000 | 6.000000e+06 | 5.340988e+06 | 7.000000 | 614.500000 | 281.000000 | 133.000000 | 1411.250000 | 0.000000 | 0.000000 | 8.599250e+03 | 65.000000 | 50.000000 | 5.800000 |
| 50% | 2005.000000 | 103.000000 | 2.350000 | 2.000000e+07 | 2.551750e+07 | 49.000000 | 988.000000 | 595.000000 | 371.500000 | 3091.000000 | 166.000000 | 1.000000 | 3.437100e+04 | 156.000000 | 110.000000 | 6.600000 |
| 75% | 2011.000000 | 118.000000 | 2.350000 | 4.500000e+07 | 6.230944e+07 | 194.750000 | 11000.000000 | 918.000000 | 636.000000 | 13758.750000 | 3000.000000 | 2.000000 | 9.634700e+04 | 326.000000 | 195.000000 | 7.200000 |
| max | 2016.000000 | 511.000000 | 16.000000 | 1.221550e+10 | 7.605058e+08 | 23000.000000 | 640000.000000 | 137000.000000 | 23000.000000 | 656730.000000 | 349000.000000 | 43.000000 | 1.689764e+06 | 5060.000000 | 813.000000 | 9.500000 |
The describe method displays below information for each of the columns.
- the count or number of values
- mean
- standard deviation
- minimum, maximum
- 25%, 50%, and 75% quantile
Please note that this information will be calculated only for the numeric values.
We can also use the corresponding method to access this information one at a time. For example, to get the mean of a particular column, you can use the mean method on that column.
movies["Gross Earnings"].mean()48468407.526809327Just like mean, there are methods available for each of the statistical information we want to access. You can read about these methods in our free pandas cheat sheet.
Reading files with no header and skipping records
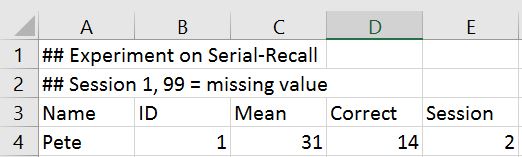
Earlier in this tutorial, we saw some ways to read a particular kind of Excel file that had headers and no rows that needed skipping. Sometimes, the Excel sheet doesn’t have any header row. For such instances, you can tell pandas not to consider the first row as header or columns names. And If the Excel sheet’s first few rows contain data that should not be read in, you can ask the read_excel method to skip a certain number of rows, starting from the top.
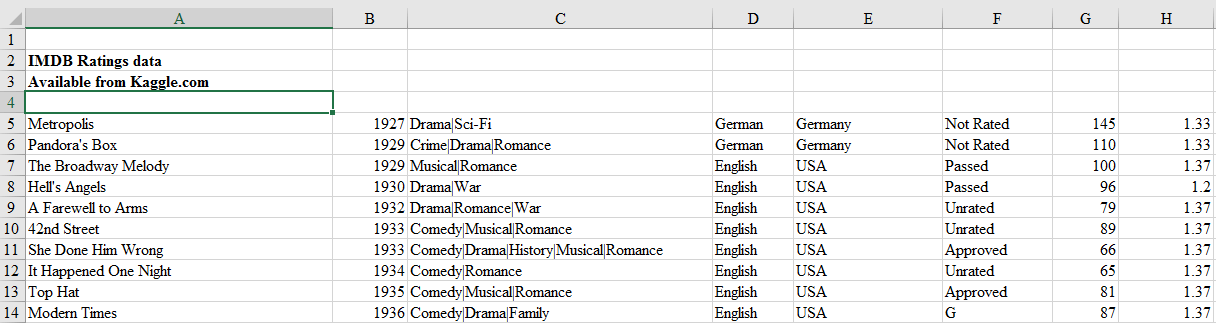
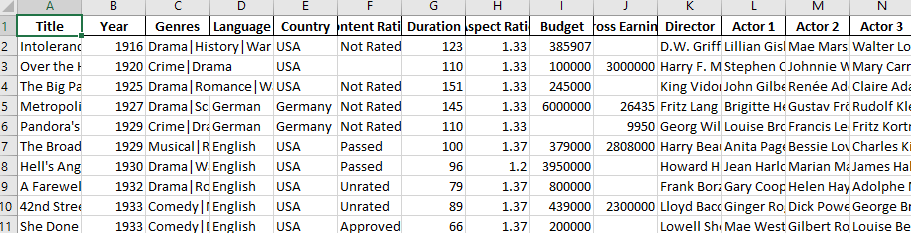
For example, look at the top few rows of this Excel file.
This file obviously has no header and first four rows are not actual records and hence should not be read in. We can tell read_excel there is no header by setting argument header to None and we can skip first four rows by setting argument skiprows to four.
movies_skip_rows = pd.read_excel("movies-no-header-skip-rows.xls", header=None, skiprows=4)
movies_skip_rows.head(5)| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | … | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Metropolis | 1927 | Drama|Sci-Fi | German | Germany | Not Rated | 145 | 1.33 | 6000000.0 | 26435.0 | … | 136 | 23 | 18.0 | 203 | 12000 | 1 | 111841 | 413 | 260.0 | 8.3 |
| 1 | Pandora’s Box | 1929 | Crime|Drama|Romance | German | Germany | Not Rated | 110 | 1.33 | NaN | 9950.0 | … | 426 | 20 | 3.0 | 455 | 926 | 1 | 7431 | 84 | 71.0 | 8.0 |
| 2 | The Broadway Melody | 1929 | Musical|Romance | English | USA | Passed | 100 | 1.37 | 379000.0 | 2808000.0 | … | 77 | 28 | 4.0 | 109 | 167 | 8 | 4546 | 71 | 36.0 | 6.3 |
| 3 | Hell’s Angels | 1930 | Drama|War | English | USA | Passed | 96 | 1.20 | 3950000.0 | NaN | … | 431 | 12 | 4.0 | 457 | 279 | 1 | 3753 | 53 | 35.0 | 7.8 |
| 4 | A Farewell to Arms | 1932 | Drama|Romance|War | English | USA | Unrated | 79 | 1.37 | 800000.0 | NaN | … | 998 | 164 | 99.0 | 1284 | 213 | 1 | 3519 | 46 | 42.0 | 6.6 |
5 rows × 25 columns
We skipped four rows from the sheet and used none of the rows as the header. Also, notice that one can combine different options in a single read statement. To skip rows at the bottom of the sheet, you can use option skip_footer, which works just like skiprows, the only difference being the rows are counted from the bottom upwards.
The column names in the previous DataFrame are numeric and were allotted as default by the pandas. We can rename the column names to descriptive ones by calling the method columns on the DataFrame and passing the column names as a list.
movies_skip_rows.columns = ['Title', 'Year', 'Genres', 'Language', 'Country', 'Content Rating', 'Duration', 'Aspect Ratio', 'Budget', 'Gross Earnings', 'Director', 'Actor 1', 'Actor 2', 'Actor 3', 'Facebook Likes - Director', 'Facebook Likes - Actor 1', 'Facebook Likes - Actor 2', 'Facebook Likes - Actor 3', 'Facebook Likes - cast Total', 'Facebook likes - Movie', 'Facenumber in posters', 'User Votes', 'Reviews by Users', 'Reviews by Crtiics', 'IMDB Score']
movies_skip_rows.head()| Title | Year | Genres | Language | Country | Content Rating | Duration | Aspect Ratio | Budget | Gross Earnings | … | Facebook Likes — Actor 1 | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Metropolis | 1927 | Drama|Sci-Fi | German | Germany | Not Rated | 145 | 1.33 | 6000000.0 | 26435.0 | … | 136 | 23 | 18.0 | 203 | 12000 | 1 | 111841 | 413 | 260.0 | 8.3 |
| 1 | Pandora’s Box | 1929 | Crime|Drama|Romance | German | Germany | Not Rated | 110 | 1.33 | NaN | 9950.0 | … | 426 | 20 | 3.0 | 455 | 926 | 1 | 7431 | 84 | 71.0 | 8.0 |
| 2 | The Broadway Melody | 1929 | Musical|Romance | English | USA | Passed | 100 | 1.37 | 379000.0 | 2808000.0 | … | 77 | 28 | 4.0 | 109 | 167 | 8 | 4546 | 71 | 36.0 | 6.3 |
| 3 | Hell’s Angels | 1930 | Drama|War | English | USA | Passed | 96 | 1.20 | 3950000.0 | NaN | … | 431 | 12 | 4.0 | 457 | 279 | 1 | 3753 | 53 | 35.0 | 7.8 |
| 4 | A Farewell to Arms | 1932 | Drama|Romance|War | English | USA | Unrated | 79 | 1.37 | 800000.0 | NaN | … | 998 | 164 | 99.0 | 1284 | 213 | 1 | 3519 | 46 | 42.0 | 6.6 |
5 rows × 25 columns
Now that we have seen how to read a subset of rows from the Excel file, we can learn how to read a subset of columns.
Reading a subset of columns
Although read_excel defaults to reading and importing all columns, you can choose to import only certain columns. By passing parse_cols=6, we are telling the read_excel method to read only the first columns till index six or first seven columns (the first column being indexed zero).
movies_subset_columns = pd.read_excel(excel_file, parse_cols=6)
movies_subset_columns.head()| Title | Year | Genres | Language | Country | Content Rating | Duration | |
|---|---|---|---|---|---|---|---|
| 0 | Intolerance: Love’s Struggle Throughout the Ages | 1916 | Drama|History|War | NaN | USA | Not Rated | 123 |
| 1 | Over the Hill to the Poorhouse | 1920 | Crime|Drama | NaN | USA | NaN | 110 |
| 2 | The Big Parade | 1925 | Drama|Romance|War | NaN | USA | Not Rated | 151 |
| 3 | Metropolis | 1927 | Drama|Sci-Fi | German | Germany | Not Rated | 145 |
| 4 | Pandora’s Box | 1929 | Crime|Drama|Romance | German | Germany | Not Rated | 110 |
Alternatively, you can pass in a list of numbers, which will let you import columns at particular indexes.
Applying formulas on the columns
One of the much-used features of Excel is to apply formulas to create new columns from existing column values. In our Excel file, we have Gross Earnings and Budget columns. We can get Net earnings by subtracting Budget from Gross earnings. We could then apply this formula in the Excel file to all the rows. We can do this in pandas also as shown below.
movies["Net Earnings"] = movies["Gross Earnings"] - movies["Budget"]Above, we used pandas to create a new column called Net Earnings, and populated it with the difference of Gross Earnings and Budget. It’s worth noting the difference here in how formulas are treated in Excel versus pandas. In Excel, a formula lives in the cell and updates when the data changes — with Python, the calculations happen and the values are stored — if Gross Earnings for one movie was manually changed, Net Earnings won’t be updated.
Let’s use the sort_values method to sort the data by the new column we created and visualize the top 10 movies by Net Earnings.
sorted_movies = movies[['Net Earnings']].sort_values(['Net Earnings'], ascending=[False])sorted_movies.head(10)['Net Earnings'].plot.barh()
plt.show()Pivot Table in pandas
Advanced Excel users also often use pivot tables. A pivot table summarizes the data of another table by grouping the data on an index and applying operations such as sorting, summing, or averaging. You can use this feature in pandas too.
We need to first identify the column or columns that will serve as the index, and the column(s) on which the summarizing formula will be applied. Let’s start small, by choosing Year as the index column and Gross Earnings as the summarization column and creating a separate DataFrame from this data.
movies_subset = movies[['Year', 'Gross Earnings']]
movies_subset.head()| Year | Gross Earnings | |
|---|---|---|
| 0 | 1916.0 | NaN |
| 1 | 1920.0 | 3000000.0 |
| 2 | 1925.0 | NaN |
| 3 | 1927.0 | 26435.0 |
| 4 | 1929.0 | 9950.0 |
We now call pivot_table on this subset of data. The method pivot_table takes a parameter index. As mentioned, we want to use Year as the index.
earnings_by_year = movies_subset.pivot_table(index=['Year'])
earnings_by_year.head()| Gross Earnings | |
|---|---|
| Year | |
| 1916.0 | NaN |
| 1920.0 | 3000000.0 |
| 1925.0 | NaN |
| 1927.0 | 26435.0 |
| 1929.0 | 1408975.0 |
This gave us a pivot table with grouping on Year and summarization on the sum of Gross Earnings. Notice, we didn’t need to specify Gross Earnings column explicitly as pandas automatically identified it the values on which summarization should be applied.
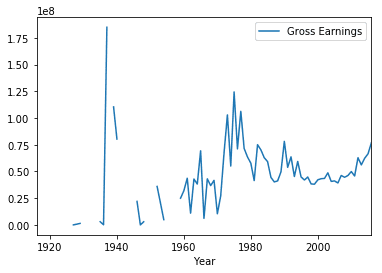
We can use this pivot table to create some data visualizations. We can call the plot method on the DataFrame to create a line plot and call the show method to display the plot in the notebook.
earnings_by_year.plot()
plt.show()We saw how to pivot with a single column as the index. Things will get more interesting if we can use multiple columns. Let’s create another DataFrame subset but this time we will choose the columns, Country, Language and Gross Earnings.
movies_subset = movies[['Country', 'Language', 'Gross Earnings']]
movies_subset.head()| Country | Language | Gross Earnings | |
|---|---|---|---|
| 0 | USA | NaN | NaN |
| 1 | USA | NaN | 3000000.0 |
| 2 | USA | NaN | NaN |
| 3 | Germany | German | 26435.0 |
| 4 | Germany | German | 9950.0 |
We will use columns Country and Language as the index for the pivot table. We will use Gross Earnings as summarization table, however, we do not need to specify this explicitly as we saw earlier.
earnings_by_co_lang = movies_subset.pivot_table(index=['Country', 'Language'])
earnings_by_co_lang.head()| Gross Earnings | ||
|---|---|---|
| Country | Language | |
| Afghanistan | Dari | 1.127331e+06 |
| Argentina | Spanish | 7.230936e+06 |
| Aruba | English | 1.007614e+07 |
| Australia | Aboriginal | 6.165429e+06 |
| Dzongkha | 5.052950e+05 |
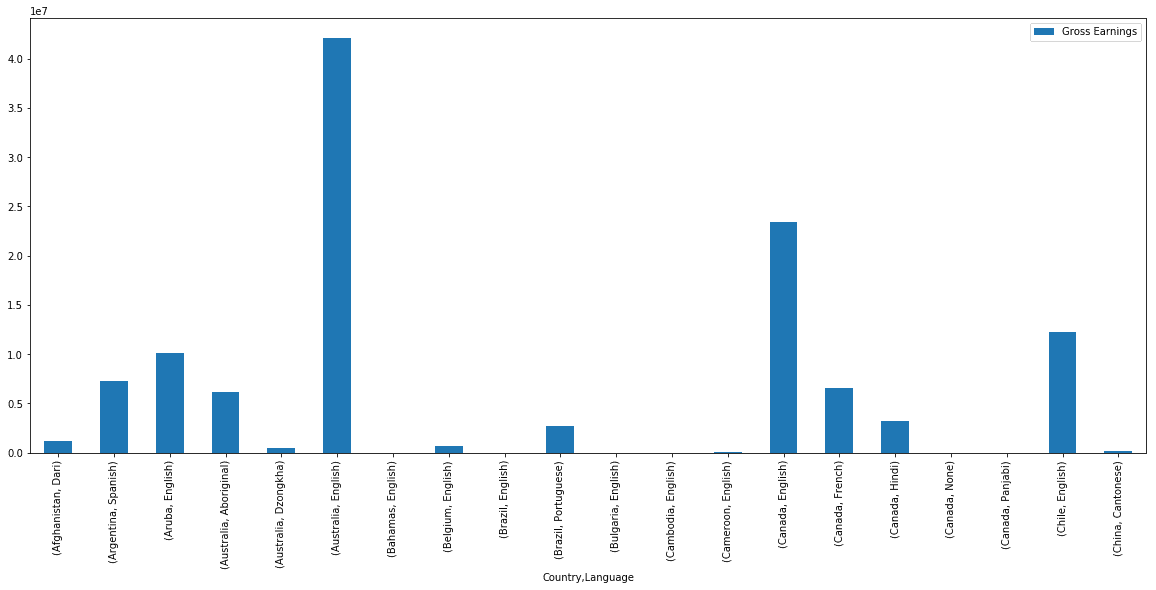
Let’s visualize this pivot table with a bar plot. Since there are still few hundred records in this pivot table, we will plot just a few of them.
earnings_by_co_lang.head(20).plot(kind='bar', figsize=(20,8))
plt.show()Exporting the results to Excel
If you’re going to be working with colleagues who use Excel, saving Excel files out of pandas is important. You can export or write a pandas DataFrame to an Excel file using pandas to_excel method. Pandas uses the xlwt Python module internally for writing to Excel files. The to_excel method is called on the DataFrame we want to export.We also need to pass a filename to which this DataFrame will be written.
movies.to_excel('output.xlsx')By default, the index is also saved to the output file. However, sometimes the index doesn’t provide any useful information. For example, the movies DataFrame has a numeric auto-increment index, that was not part of the original Excel data.
movies.head()| Title | Year | Genres | Language | Country | Content Rating | Duration | Aspect Ratio | Budget | Gross Earnings | … | Facebook Likes — Actor 2 | Facebook Likes — Actor 3 | Facebook Likes — cast Total | Facebook likes — Movie | Facenumber in posters | User Votes | Reviews by Users | Reviews by Crtiics | IMDB Score | Net Earnings | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Intolerance: Love’s Struggle Throughout the Ages | 1916.0 | Drama|History|War | NaN | USA | Not Rated | 123.0 | 1.33 | 385907.0 | NaN | … | 22.0 | 9.0 | 481 | 691 | 1.0 | 10718 | 88.0 | 69.0 | 8.0 | NaN |
| 1 | Over the Hill to the Poorhouse | 1920.0 | Crime|Drama | NaN | USA | NaN | 110.0 | 1.33 | 100000.0 | 3000000.0 | … | 2.0 | 0.0 | 4 | 0 | 1.0 | 5 | 1.0 | 1.0 | 4.8 | 2900000.0 |
| 2 | The Big Parade | 1925.0 | Drama|Romance|War | NaN | USA | Not Rated | 151.0 | 1.33 | 245000.0 | NaN | … | 12.0 | 6.0 | 108 | 226 | 0.0 | 4849 | 45.0 | 48.0 | 8.3 | NaN |
| 3 | Metropolis | 1927.0 | Drama|Sci-Fi | German | Germany | Not Rated | 145.0 | 1.33 | 6000000.0 | 26435.0 | … | 23.0 | 18.0 | 203 | 12000 | 1.0 | 111841 | 413.0 | 260.0 | 8.3 | -5973565.0 |
| 4 | Pandora’s Box | 1929.0 | Crime|Drama|Romance | German | Germany | Not Rated | 110.0 | 1.33 | NaN | 9950.0 | … | 20.0 | 3.0 | 455 | 926 | 1.0 | 7431 | 84.0 | 71.0 | 8.0 | NaN |
5 rows × 26 columns
You can choose to skip the index by passing along index-False.
movies.to_excel('output.xlsx', index=False)We need to be able to make our output files look nice before we can send it out to our co-workers. We can use pandas ExcelWriter class along with the XlsxWriter Python module to apply the formatting.
We can do use these advanced output options by creating a ExcelWriter object and use this object to write to the EXcel file.
writer = pd.ExcelWriter('output.xlsx', engine='xlsxwriter')
movies.to_excel(writer, index=False, sheet_name='report')
workbook = writer.bookworksheet = writer.sheets['report']We can apply customizations by calling add_format on the workbook we are writing to. Here we are setting header format as bold.
header_fmt = workbook.add_format({'bold': True})
worksheet.set_row(0, None, header_fmt)Finally, we save the output file by calling the method save on the writer object.
writer.save()As an example, we saved the data with column headers set as bold. And the saved file looks like the image below.
Like this, one can use XlsxWriter to apply various formatting to the output Excel file.
Conclusion
Pandas is not a replacement for Excel. Both tools have their place in the data analysis workflow and can be very great companion tools. As we demonstrated, pandas can do a lot of complex data analysis and manipulations, which depending on your need and expertise, can go beyond what you can achieve if you are just using Excel. One of the major benefits of using Python and pandas over Excel is that it helps you automate Excel file processing by writing scripts and integrating with your automated data workflow. Pandas also has excellent methods for reading all kinds of data from Excel files. You can export your results from pandas back to Excel too if that’s preferred by your intended audience.
On the other hand, Excel is a such a widely used data tool, it’s not a wise to ignore it. Acquiring expertise in both pandas and Excel and making them work together gives you skills that can help you stand out in your organization.
If you’d like to learn more about this topic, check out Dataquest’s interactive Pandas and NumPy Fundamentals course, and our Data Analyst in Python, and Data Scientist in Python paths that will help you become job-ready in around 6 months.
Watch Now This tutorial has a related video course created by the Real Python team. Watch it together with the written tutorial to deepen your understanding: Reading and Writing Files With Pandas
pandas is a powerful and flexible Python package that allows you to work with labeled and time series data. It also provides statistics methods, enables plotting, and more. One crucial feature of pandas is its ability to write and read Excel, CSV, and many other types of files. Functions like the pandas read_csv() method enable you to work with files effectively. You can use them to save the data and labels from pandas objects to a file and load them later as pandas Series or DataFrame instances.
In this tutorial, you’ll learn:
- What the pandas IO tools API is
- How to read and write data to and from files
- How to work with various file formats
- How to work with big data efficiently
Let’s start reading and writing files!
Installing pandas
The code in this tutorial is executed with CPython 3.7.4 and pandas 0.25.1. It would be beneficial to make sure you have the latest versions of Python and pandas on your machine. You might want to create a new virtual environment and install the dependencies for this tutorial.
First, you’ll need the pandas library. You may already have it installed. If you don’t, then you can install it with pip:
Once the installation process completes, you should have pandas installed and ready.
Anaconda is an excellent Python distribution that comes with Python, many useful packages like pandas, and a package and environment manager called Conda. To learn more about Anaconda, check out Setting Up Python for Machine Learning on Windows.
If you don’t have pandas in your virtual environment, then you can install it with Conda:
Conda is powerful as it manages the dependencies and their versions. To learn more about working with Conda, you can check out the official documentation.
Preparing Data
In this tutorial, you’ll use the data related to 20 countries. Here’s an overview of the data and sources you’ll be working with:
-
Country is denoted by the country name. Each country is in the top 10 list for either population, area, or gross domestic product (GDP). The row labels for the dataset are the three-letter country codes defined in ISO 3166-1. The column label for the dataset is
COUNTRY. -
Population is expressed in millions. The data comes from a list of countries and dependencies by population on Wikipedia. The column label for the dataset is
POP. -
Area is expressed in thousands of kilometers squared. The data comes from a list of countries and dependencies by area on Wikipedia. The column label for the dataset is
AREA. -
Gross domestic product is expressed in millions of U.S. dollars, according to the United Nations data for 2017. You can find this data in the list of countries by nominal GDP on Wikipedia. The column label for the dataset is
GDP. -
Continent is either Africa, Asia, Oceania, Europe, North America, or South America. You can find this information on Wikipedia as well. The column label for the dataset is
CONT. -
Independence day is a date that commemorates a nation’s independence. The data comes from the list of national independence days on Wikipedia. The dates are shown in ISO 8601 format. The first four digits represent the year, the next two numbers are the month, and the last two are for the day of the month. The column label for the dataset is
IND_DAY.
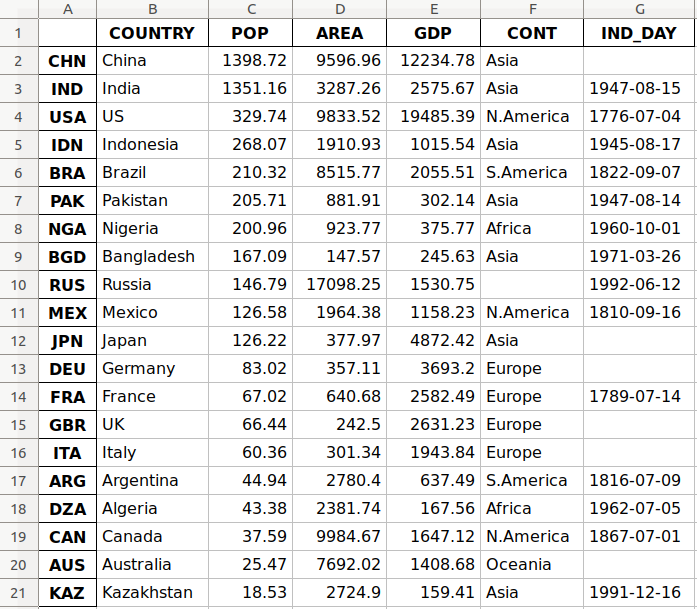
This is how the data looks as a table:
| COUNTRY | POP | AREA | GDP | CONT | IND_DAY | |
|---|---|---|---|---|---|---|
| CHN | China | 1398.72 | 9596.96 | 12234.78 | Asia | |
| IND | India | 1351.16 | 3287.26 | 2575.67 | Asia | 1947-08-15 |
| USA | US | 329.74 | 9833.52 | 19485.39 | N.America | 1776-07-04 |
| IDN | Indonesia | 268.07 | 1910.93 | 1015.54 | Asia | 1945-08-17 |
| BRA | Brazil | 210.32 | 8515.77 | 2055.51 | S.America | 1822-09-07 |
| PAK | Pakistan | 205.71 | 881.91 | 302.14 | Asia | 1947-08-14 |
| NGA | Nigeria | 200.96 | 923.77 | 375.77 | Africa | 1960-10-01 |
| BGD | Bangladesh | 167.09 | 147.57 | 245.63 | Asia | 1971-03-26 |
| RUS | Russia | 146.79 | 17098.25 | 1530.75 | 1992-06-12 | |
| MEX | Mexico | 126.58 | 1964.38 | 1158.23 | N.America | 1810-09-16 |
| JPN | Japan | 126.22 | 377.97 | 4872.42 | Asia | |
| DEU | Germany | 83.02 | 357.11 | 3693.20 | Europe | |
| FRA | France | 67.02 | 640.68 | 2582.49 | Europe | 1789-07-14 |
| GBR | UK | 66.44 | 242.50 | 2631.23 | Europe | |
| ITA | Italy | 60.36 | 301.34 | 1943.84 | Europe | |
| ARG | Argentina | 44.94 | 2780.40 | 637.49 | S.America | 1816-07-09 |
| DZA | Algeria | 43.38 | 2381.74 | 167.56 | Africa | 1962-07-05 |
| CAN | Canada | 37.59 | 9984.67 | 1647.12 | N.America | 1867-07-01 |
| AUS | Australia | 25.47 | 7692.02 | 1408.68 | Oceania | |
| KAZ | Kazakhstan | 18.53 | 2724.90 | 159.41 | Asia | 1991-12-16 |
You may notice that some of the data is missing. For example, the continent for Russia is not specified because it spreads across both Europe and Asia. There are also several missing independence days because the data source omits them.
You can organize this data in Python using a nested dictionary:
data = {
'CHN': {'COUNTRY': 'China', 'POP': 1_398.72, 'AREA': 9_596.96,
'GDP': 12_234.78, 'CONT': 'Asia'},
'IND': {'COUNTRY': 'India', 'POP': 1_351.16, 'AREA': 3_287.26,
'GDP': 2_575.67, 'CONT': 'Asia', 'IND_DAY': '1947-08-15'},
'USA': {'COUNTRY': 'US', 'POP': 329.74, 'AREA': 9_833.52,
'GDP': 19_485.39, 'CONT': 'N.America',
'IND_DAY': '1776-07-04'},
'IDN': {'COUNTRY': 'Indonesia', 'POP': 268.07, 'AREA': 1_910.93,
'GDP': 1_015.54, 'CONT': 'Asia', 'IND_DAY': '1945-08-17'},
'BRA': {'COUNTRY': 'Brazil', 'POP': 210.32, 'AREA': 8_515.77,
'GDP': 2_055.51, 'CONT': 'S.America', 'IND_DAY': '1822-09-07'},
'PAK': {'COUNTRY': 'Pakistan', 'POP': 205.71, 'AREA': 881.91,
'GDP': 302.14, 'CONT': 'Asia', 'IND_DAY': '1947-08-14'},
'NGA': {'COUNTRY': 'Nigeria', 'POP': 200.96, 'AREA': 923.77,
'GDP': 375.77, 'CONT': 'Africa', 'IND_DAY': '1960-10-01'},
'BGD': {'COUNTRY': 'Bangladesh', 'POP': 167.09, 'AREA': 147.57,
'GDP': 245.63, 'CONT': 'Asia', 'IND_DAY': '1971-03-26'},
'RUS': {'COUNTRY': 'Russia', 'POP': 146.79, 'AREA': 17_098.25,
'GDP': 1_530.75, 'IND_DAY': '1992-06-12'},
'MEX': {'COUNTRY': 'Mexico', 'POP': 126.58, 'AREA': 1_964.38,
'GDP': 1_158.23, 'CONT': 'N.America', 'IND_DAY': '1810-09-16'},
'JPN': {'COUNTRY': 'Japan', 'POP': 126.22, 'AREA': 377.97,
'GDP': 4_872.42, 'CONT': 'Asia'},
'DEU': {'COUNTRY': 'Germany', 'POP': 83.02, 'AREA': 357.11,
'GDP': 3_693.20, 'CONT': 'Europe'},
'FRA': {'COUNTRY': 'France', 'POP': 67.02, 'AREA': 640.68,
'GDP': 2_582.49, 'CONT': 'Europe', 'IND_DAY': '1789-07-14'},
'GBR': {'COUNTRY': 'UK', 'POP': 66.44, 'AREA': 242.50,
'GDP': 2_631.23, 'CONT': 'Europe'},
'ITA': {'COUNTRY': 'Italy', 'POP': 60.36, 'AREA': 301.34,
'GDP': 1_943.84, 'CONT': 'Europe'},
'ARG': {'COUNTRY': 'Argentina', 'POP': 44.94, 'AREA': 2_780.40,
'GDP': 637.49, 'CONT': 'S.America', 'IND_DAY': '1816-07-09'},
'DZA': {'COUNTRY': 'Algeria', 'POP': 43.38, 'AREA': 2_381.74,
'GDP': 167.56, 'CONT': 'Africa', 'IND_DAY': '1962-07-05'},
'CAN': {'COUNTRY': 'Canada', 'POP': 37.59, 'AREA': 9_984.67,
'GDP': 1_647.12, 'CONT': 'N.America', 'IND_DAY': '1867-07-01'},
'AUS': {'COUNTRY': 'Australia', 'POP': 25.47, 'AREA': 7_692.02,
'GDP': 1_408.68, 'CONT': 'Oceania'},
'KAZ': {'COUNTRY': 'Kazakhstan', 'POP': 18.53, 'AREA': 2_724.90,
'GDP': 159.41, 'CONT': 'Asia', 'IND_DAY': '1991-12-16'}
}
columns = ('COUNTRY', 'POP', 'AREA', 'GDP', 'CONT', 'IND_DAY')
Each row of the table is written as an inner dictionary whose keys are the column names and values are the corresponding data. These dictionaries are then collected as the values in the outer data dictionary. The corresponding keys for data are the three-letter country codes.
You can use this data to create an instance of a pandas DataFrame. First, you need to import pandas:
>>>
>>> import pandas as pd
Now that you have pandas imported, you can use the DataFrame constructor and data to create a DataFrame object.
data is organized in such a way that the country codes correspond to columns. You can reverse the rows and columns of a DataFrame with the property .T:
>>>
>>> df = pd.DataFrame(data=data).T
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.8 Asia NaN
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.4 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.2 1530.75 NaN 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaN
DEU Germany 83.02 357.11 3693.2 Europe NaN
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.5 2631.23 Europe NaN
ITA Italy 60.36 301.34 1943.84 Europe NaN
ARG Argentina 44.94 2780.4 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaN
KAZ Kazakhstan 18.53 2724.9 159.41 Asia 1991-12-16
Now you have your DataFrame object populated with the data about each country.
Versions of Python older than 3.6 did not guarantee the order of keys in dictionaries. To ensure the order of columns is maintained for older versions of Python and pandas, you can specify index=columns:
>>>
>>> df = pd.DataFrame(data=data, index=columns).T
Now that you’ve prepared your data, you’re ready to start working with files!
Using the pandas read_csv() and .to_csv() Functions
A comma-separated values (CSV) file is a plaintext file with a .csv extension that holds tabular data. This is one of the most popular file formats for storing large amounts of data. Each row of the CSV file represents a single table row. The values in the same row are by default separated with commas, but you could change the separator to a semicolon, tab, space, or some other character.
Write a CSV File
You can save your pandas DataFrame as a CSV file with .to_csv():
>>>
>>> df.to_csv('data.csv')
That’s it! You’ve created the file data.csv in your current working directory. You can expand the code block below to see how your CSV file should look:
,COUNTRY,POP,AREA,GDP,CONT,IND_DAY
CHN,China,1398.72,9596.96,12234.78,Asia,
IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15
USA,US,329.74,9833.52,19485.39,N.America,1776-07-04
IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17
BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07
PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14
NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01
BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26
RUS,Russia,146.79,17098.25,1530.75,,1992-06-12
MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16
JPN,Japan,126.22,377.97,4872.42,Asia,
DEU,Germany,83.02,357.11,3693.2,Europe,
FRA,France,67.02,640.68,2582.49,Europe,1789-07-14
GBR,UK,66.44,242.5,2631.23,Europe,
ITA,Italy,60.36,301.34,1943.84,Europe,
ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09
DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05
CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01
AUS,Australia,25.47,7692.02,1408.68,Oceania,
KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
This text file contains the data separated with commas. The first column contains the row labels. In some cases, you’ll find them irrelevant. If you don’t want to keep them, then you can pass the argument index=False to .to_csv().
Read a CSV File
Once your data is saved in a CSV file, you’ll likely want to load and use it from time to time. You can do that with the pandas read_csv() function:
>>>
>>> df = pd.read_csv('data.csv', index_col=0)
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.78 Asia NaN
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaN
DEU Germany 83.02 357.11 3693.20 Europe NaN
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaN
ITA Italy 60.36 301.34 1943.84 Europe NaN
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaN
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
In this case, the pandas read_csv() function returns a new DataFrame with the data and labels from the file data.csv, which you specified with the first argument. This string can be any valid path, including URLs.
The parameter index_col specifies the column from the CSV file that contains the row labels. You assign a zero-based column index to this parameter. You should determine the value of index_col when the CSV file contains the row labels to avoid loading them as data.
You’ll learn more about using pandas with CSV files later on in this tutorial. You can also check out Reading and Writing CSV Files in Python to see how to handle CSV files with the built-in Python library csv as well.
Using pandas to Write and Read Excel Files
Microsoft Excel is probably the most widely-used spreadsheet software. While older versions used binary .xls files, Excel 2007 introduced the new XML-based .xlsx file. You can read and write Excel files in pandas, similar to CSV files. However, you’ll need to install the following Python packages first:
- xlwt to write to
.xlsfiles - openpyxl or XlsxWriter to write to
.xlsxfiles - xlrd to read Excel files
You can install them using pip with a single command:
$ pip install xlwt openpyxl xlsxwriter xlrd
You can also use Conda:
$ conda install xlwt openpyxl xlsxwriter xlrd
Please note that you don’t have to install all these packages. For example, you don’t need both openpyxl and XlsxWriter. If you’re going to work just with .xls files, then you don’t need any of them! However, if you intend to work only with .xlsx files, then you’re going to need at least one of them, but not xlwt. Take some time to decide which packages are right for your project.
Write an Excel File
Once you have those packages installed, you can save your DataFrame in an Excel file with .to_excel():
>>>
>>> df.to_excel('data.xlsx')
The argument 'data.xlsx' represents the target file and, optionally, its path. The above statement should create the file data.xlsx in your current working directory. That file should look like this:
The first column of the file contains the labels of the rows, while the other columns store data.
Read an Excel File
You can load data from Excel files with read_excel():
>>>
>>> df = pd.read_excel('data.xlsx', index_col=0)
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.78 Asia NaN
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaN
DEU Germany 83.02 357.11 3693.20 Europe NaN
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaN
ITA Italy 60.36 301.34 1943.84 Europe NaN
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaN
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
read_excel() returns a new DataFrame that contains the values from data.xlsx. You can also use read_excel() with OpenDocument spreadsheets, or .ods files.
You’ll learn more about working with Excel files later on in this tutorial. You can also check out Using pandas to Read Large Excel Files in Python.
Understanding the pandas IO API
pandas IO Tools is the API that allows you to save the contents of Series and DataFrame objects to the clipboard, objects, or files of various types. It also enables loading data from the clipboard, objects, or files.
Write Files
Series and DataFrame objects have methods that enable writing data and labels to the clipboard or files. They’re named with the pattern .to_<file-type>(), where <file-type> is the type of the target file.
You’ve learned about .to_csv() and .to_excel(), but there are others, including:
.to_json().to_html().to_sql().to_pickle()
There are still more file types that you can write to, so this list is not exhaustive.
These methods have parameters specifying the target file path where you saved the data and labels. This is mandatory in some cases and optional in others. If this option is available and you choose to omit it, then the methods return the objects (like strings or iterables) with the contents of DataFrame instances.
The optional parameter compression decides how to compress the file with the data and labels. You’ll learn more about it later on. There are a few other parameters, but they’re mostly specific to one or several methods. You won’t go into them in detail here.
Read Files
pandas functions for reading the contents of files are named using the pattern .read_<file-type>(), where <file-type> indicates the type of the file to read. You’ve already seen the pandas read_csv() and read_excel() functions. Here are a few others:
read_json()read_html()read_sql()read_pickle()
These functions have a parameter that specifies the target file path. It can be any valid string that represents the path, either on a local machine or in a URL. Other objects are also acceptable depending on the file type.
The optional parameter compression determines the type of decompression to use for the compressed files. You’ll learn about it later on in this tutorial. There are other parameters, but they’re specific to one or several functions. You won’t go into them in detail here.
Working With Different File Types
The pandas library offers a wide range of possibilities for saving your data to files and loading data from files. In this section, you’ll learn more about working with CSV and Excel files. You’ll also see how to use other types of files, like JSON, web pages, databases, and Python pickle files.
CSV Files
You’ve already learned how to read and write CSV files. Now let’s dig a little deeper into the details. When you use .to_csv() to save your DataFrame, you can provide an argument for the parameter path_or_buf to specify the path, name, and extension of the target file.
path_or_buf is the first argument .to_csv() will get. It can be any string that represents a valid file path that includes the file name and its extension. You’ve seen this in a previous example. However, if you omit path_or_buf, then .to_csv() won’t create any files. Instead, it’ll return the corresponding string:
>>>
>>> df = pd.DataFrame(data=data).T
>>> s = df.to_csv()
>>> print(s)
,COUNTRY,POP,AREA,GDP,CONT,IND_DAY
CHN,China,1398.72,9596.96,12234.78,Asia,
IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15
USA,US,329.74,9833.52,19485.39,N.America,1776-07-04
IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17
BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07
PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14
NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01
BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26
RUS,Russia,146.79,17098.25,1530.75,,1992-06-12
MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16
JPN,Japan,126.22,377.97,4872.42,Asia,
DEU,Germany,83.02,357.11,3693.2,Europe,
FRA,France,67.02,640.68,2582.49,Europe,1789-07-14
GBR,UK,66.44,242.5,2631.23,Europe,
ITA,Italy,60.36,301.34,1943.84,Europe,
ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09
DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05
CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01
AUS,Australia,25.47,7692.02,1408.68,Oceania,
KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
Now you have the string s instead of a CSV file. You also have some missing values in your DataFrame object. For example, the continent for Russia and the independence days for several countries (China, Japan, and so on) are not available. In data science and machine learning, you must handle missing values carefully. pandas excels here! By default, pandas uses the NaN value to replace the missing values.
The continent that corresponds to Russia in df is nan:
>>>
>>> df.loc['RUS', 'CONT']
nan
This example uses .loc[] to get data with the specified row and column names.
When you save your DataFrame to a CSV file, empty strings ('') will represent the missing data. You can see this both in your file data.csv and in the string s. If you want to change this behavior, then use the optional parameter na_rep:
>>>
>>> df.to_csv('new-data.csv', na_rep='(missing)')
This code produces the file new-data.csv where the missing values are no longer empty strings. You can expand the code block below to see how this file should look:
,COUNTRY,POP,AREA,GDP,CONT,IND_DAY
CHN,China,1398.72,9596.96,12234.78,Asia,(missing)
IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15
USA,US,329.74,9833.52,19485.39,N.America,1776-07-04
IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17
BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07
PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14
NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01
BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26
RUS,Russia,146.79,17098.25,1530.75,(missing),1992-06-12
MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16
JPN,Japan,126.22,377.97,4872.42,Asia,(missing)
DEU,Germany,83.02,357.11,3693.2,Europe,(missing)
FRA,France,67.02,640.68,2582.49,Europe,1789-07-14
GBR,UK,66.44,242.5,2631.23,Europe,(missing)
ITA,Italy,60.36,301.34,1943.84,Europe,(missing)
ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09
DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05
CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01
AUS,Australia,25.47,7692.02,1408.68,Oceania,(missing)
KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
Now, the string '(missing)' in the file corresponds to the nan values from df.
When pandas reads files, it considers the empty string ('') and a few others as missing values by default:
'nan''-nan''NA''N/A''NaN''null'
If you don’t want this behavior, then you can pass keep_default_na=False to the pandas read_csv() function. To specify other labels for missing values, use the parameter na_values:
>>>
>>> pd.read_csv('new-data.csv', index_col=0, na_values='(missing)')
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.78 Asia NaN
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaN
DEU Germany 83.02 357.11 3693.20 Europe NaN
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaN
ITA Italy 60.36 301.34 1943.84 Europe NaN
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaN
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
Here, you’ve marked the string '(missing)' as a new missing data label, and pandas replaced it with nan when it read the file.
When you load data from a file, pandas assigns the data types to the values of each column by default. You can check these types with .dtypes:
>>>
>>> df = pd.read_csv('data.csv', index_col=0)
>>> df.dtypes
COUNTRY object
POP float64
AREA float64
GDP float64
CONT object
IND_DAY object
dtype: object
The columns with strings and dates ('COUNTRY', 'CONT', and 'IND_DAY') have the data type object. Meanwhile, the numeric columns contain 64-bit floating-point numbers (float64).
You can use the parameter dtype to specify the desired data types and parse_dates to force use of datetimes:
>>>
>>> dtypes = {'POP': 'float32', 'AREA': 'float32', 'GDP': 'float32'}
>>> df = pd.read_csv('data.csv', index_col=0, dtype=dtypes,
... parse_dates=['IND_DAY'])
>>> df.dtypes
COUNTRY object
POP float32
AREA float32
GDP float32
CONT object
IND_DAY datetime64[ns]
dtype: object
>>> df['IND_DAY']
CHN NaT
IND 1947-08-15
USA 1776-07-04
IDN 1945-08-17
BRA 1822-09-07
PAK 1947-08-14
NGA 1960-10-01
BGD 1971-03-26
RUS 1992-06-12
MEX 1810-09-16
JPN NaT
DEU NaT
FRA 1789-07-14
GBR NaT
ITA NaT
ARG 1816-07-09
DZA 1962-07-05
CAN 1867-07-01
AUS NaT
KAZ 1991-12-16
Name: IND_DAY, dtype: datetime64[ns]
Now, you have 32-bit floating-point numbers (float32) as specified with dtype. These differ slightly from the original 64-bit numbers because of smaller precision. The values in the last column are considered as dates and have the data type datetime64. That’s why the NaN values in this column are replaced with NaT.
Now that you have real dates, you can save them in the format you like:
>>>
>>> df = pd.read_csv('data.csv', index_col=0, parse_dates=['IND_DAY'])
>>> df.to_csv('formatted-data.csv', date_format='%B %d, %Y')
Here, you’ve specified the parameter date_format to be '%B %d, %Y'. You can expand the code block below to see the resulting file:
,COUNTRY,POP,AREA,GDP,CONT,IND_DAY
CHN,China,1398.72,9596.96,12234.78,Asia,
IND,India,1351.16,3287.26,2575.67,Asia,"August 15, 1947"
USA,US,329.74,9833.52,19485.39,N.America,"July 04, 1776"
IDN,Indonesia,268.07,1910.93,1015.54,Asia,"August 17, 1945"
BRA,Brazil,210.32,8515.77,2055.51,S.America,"September 07, 1822"
PAK,Pakistan,205.71,881.91,302.14,Asia,"August 14, 1947"
NGA,Nigeria,200.96,923.77,375.77,Africa,"October 01, 1960"
BGD,Bangladesh,167.09,147.57,245.63,Asia,"March 26, 1971"
RUS,Russia,146.79,17098.25,1530.75,,"June 12, 1992"
MEX,Mexico,126.58,1964.38,1158.23,N.America,"September 16, 1810"
JPN,Japan,126.22,377.97,4872.42,Asia,
DEU,Germany,83.02,357.11,3693.2,Europe,
FRA,France,67.02,640.68,2582.49,Europe,"July 14, 1789"
GBR,UK,66.44,242.5,2631.23,Europe,
ITA,Italy,60.36,301.34,1943.84,Europe,
ARG,Argentina,44.94,2780.4,637.49,S.America,"July 09, 1816"
DZA,Algeria,43.38,2381.74,167.56,Africa,"July 05, 1962"
CAN,Canada,37.59,9984.67,1647.12,N.America,"July 01, 1867"
AUS,Australia,25.47,7692.02,1408.68,Oceania,
KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,"December 16, 1991"
The format of the dates is different now. The format '%B %d, %Y' means the date will first display the full name of the month, then the day followed by a comma, and finally the full year.
There are several other optional parameters that you can use with .to_csv():
sepdenotes a values separator.decimalindicates a decimal separator.encodingsets the file encoding.headerspecifies whether you want to write column labels in the file.
Here’s how you would pass arguments for sep and header:
>>>
>>> s = df.to_csv(sep=';', header=False)
>>> print(s)
CHN;China;1398.72;9596.96;12234.78;Asia;
IND;India;1351.16;3287.26;2575.67;Asia;1947-08-15
USA;US;329.74;9833.52;19485.39;N.America;1776-07-04
IDN;Indonesia;268.07;1910.93;1015.54;Asia;1945-08-17
BRA;Brazil;210.32;8515.77;2055.51;S.America;1822-09-07
PAK;Pakistan;205.71;881.91;302.14;Asia;1947-08-14
NGA;Nigeria;200.96;923.77;375.77;Africa;1960-10-01
BGD;Bangladesh;167.09;147.57;245.63;Asia;1971-03-26
RUS;Russia;146.79;17098.25;1530.75;;1992-06-12
MEX;Mexico;126.58;1964.38;1158.23;N.America;1810-09-16
JPN;Japan;126.22;377.97;4872.42;Asia;
DEU;Germany;83.02;357.11;3693.2;Europe;
FRA;France;67.02;640.68;2582.49;Europe;1789-07-14
GBR;UK;66.44;242.5;2631.23;Europe;
ITA;Italy;60.36;301.34;1943.84;Europe;
ARG;Argentina;44.94;2780.4;637.49;S.America;1816-07-09
DZA;Algeria;43.38;2381.74;167.56;Africa;1962-07-05
CAN;Canada;37.59;9984.67;1647.12;N.America;1867-07-01
AUS;Australia;25.47;7692.02;1408.68;Oceania;
KAZ;Kazakhstan;18.53;2724.9;159.41;Asia;1991-12-16
The data is separated with a semicolon (';') because you’ve specified sep=';'. Also, since you passed header=False, you see your data without the header row of column names.
The pandas read_csv() function has many additional options for managing missing data, working with dates and times, quoting, encoding, handling errors, and more. For instance, if you have a file with one data column and want to get a Series object instead of a DataFrame, then you can pass squeeze=True to read_csv(). You’ll learn later on about data compression and decompression, as well as how to skip rows and columns.
JSON Files
JSON stands for JavaScript object notation. JSON files are plaintext files used for data interchange, and humans can read them easily. They follow the ISO/IEC 21778:2017 and ECMA-404 standards and use the .json extension. Python and pandas work well with JSON files, as Python’s json library offers built-in support for them.
You can save the data from your DataFrame to a JSON file with .to_json(). Start by creating a DataFrame object again. Use the dictionary data that holds the data about countries and then apply .to_json():
>>>
>>> df = pd.DataFrame(data=data).T
>>> df.to_json('data-columns.json')
This code produces the file data-columns.json. You can expand the code block below to see how this file should look:
{"COUNTRY":{"CHN":"China","IND":"India","USA":"US","IDN":"Indonesia","BRA":"Brazil","PAK":"Pakistan","NGA":"Nigeria","BGD":"Bangladesh","RUS":"Russia","MEX":"Mexico","JPN":"Japan","DEU":"Germany","FRA":"France","GBR":"UK","ITA":"Italy","ARG":"Argentina","DZA":"Algeria","CAN":"Canada","AUS":"Australia","KAZ":"Kazakhstan"},"POP":{"CHN":1398.72,"IND":1351.16,"USA":329.74,"IDN":268.07,"BRA":210.32,"PAK":205.71,"NGA":200.96,"BGD":167.09,"RUS":146.79,"MEX":126.58,"JPN":126.22,"DEU":83.02,"FRA":67.02,"GBR":66.44,"ITA":60.36,"ARG":44.94,"DZA":43.38,"CAN":37.59,"AUS":25.47,"KAZ":18.53},"AREA":{"CHN":9596.96,"IND":3287.26,"USA":9833.52,"IDN":1910.93,"BRA":8515.77,"PAK":881.91,"NGA":923.77,"BGD":147.57,"RUS":17098.25,"MEX":1964.38,"JPN":377.97,"DEU":357.11,"FRA":640.68,"GBR":242.5,"ITA":301.34,"ARG":2780.4,"DZA":2381.74,"CAN":9984.67,"AUS":7692.02,"KAZ":2724.9},"GDP":{"CHN":12234.78,"IND":2575.67,"USA":19485.39,"IDN":1015.54,"BRA":2055.51,"PAK":302.14,"NGA":375.77,"BGD":245.63,"RUS":1530.75,"MEX":1158.23,"JPN":4872.42,"DEU":3693.2,"FRA":2582.49,"GBR":2631.23,"ITA":1943.84,"ARG":637.49,"DZA":167.56,"CAN":1647.12,"AUS":1408.68,"KAZ":159.41},"CONT":{"CHN":"Asia","IND":"Asia","USA":"N.America","IDN":"Asia","BRA":"S.America","PAK":"Asia","NGA":"Africa","BGD":"Asia","RUS":null,"MEX":"N.America","JPN":"Asia","DEU":"Europe","FRA":"Europe","GBR":"Europe","ITA":"Europe","ARG":"S.America","DZA":"Africa","CAN":"N.America","AUS":"Oceania","KAZ":"Asia"},"IND_DAY":{"CHN":null,"IND":"1947-08-15","USA":"1776-07-04","IDN":"1945-08-17","BRA":"1822-09-07","PAK":"1947-08-14","NGA":"1960-10-01","BGD":"1971-03-26","RUS":"1992-06-12","MEX":"1810-09-16","JPN":null,"DEU":null,"FRA":"1789-07-14","GBR":null,"ITA":null,"ARG":"1816-07-09","DZA":"1962-07-05","CAN":"1867-07-01","AUS":null,"KAZ":"1991-12-16"}}
data-columns.json has one large dictionary with the column labels as keys and the corresponding inner dictionaries as values.
You can get a different file structure if you pass an argument for the optional parameter orient:
>>>
>>> df.to_json('data-index.json', orient='index')
The orient parameter defaults to 'columns'. Here, you’ve set it to index.
You should get a new file data-index.json. You can expand the code block below to see the changes:
{"CHN":{"COUNTRY":"China","POP":1398.72,"AREA":9596.96,"GDP":12234.78,"CONT":"Asia","IND_DAY":null},"IND":{"COUNTRY":"India","POP":1351.16,"AREA":3287.26,"GDP":2575.67,"CONT":"Asia","IND_DAY":"1947-08-15"},"USA":{"COUNTRY":"US","POP":329.74,"AREA":9833.52,"GDP":19485.39,"CONT":"N.America","IND_DAY":"1776-07-04"},"IDN":{"COUNTRY":"Indonesia","POP":268.07,"AREA":1910.93,"GDP":1015.54,"CONT":"Asia","IND_DAY":"1945-08-17"},"BRA":{"COUNTRY":"Brazil","POP":210.32,"AREA":8515.77,"GDP":2055.51,"CONT":"S.America","IND_DAY":"1822-09-07"},"PAK":{"COUNTRY":"Pakistan","POP":205.71,"AREA":881.91,"GDP":302.14,"CONT":"Asia","IND_DAY":"1947-08-14"},"NGA":{"COUNTRY":"Nigeria","POP":200.96,"AREA":923.77,"GDP":375.77,"CONT":"Africa","IND_DAY":"1960-10-01"},"BGD":{"COUNTRY":"Bangladesh","POP":167.09,"AREA":147.57,"GDP":245.63,"CONT":"Asia","IND_DAY":"1971-03-26"},"RUS":{"COUNTRY":"Russia","POP":146.79,"AREA":17098.25,"GDP":1530.75,"CONT":null,"IND_DAY":"1992-06-12"},"MEX":{"COUNTRY":"Mexico","POP":126.58,"AREA":1964.38,"GDP":1158.23,"CONT":"N.America","IND_DAY":"1810-09-16"},"JPN":{"COUNTRY":"Japan","POP":126.22,"AREA":377.97,"GDP":4872.42,"CONT":"Asia","IND_DAY":null},"DEU":{"COUNTRY":"Germany","POP":83.02,"AREA":357.11,"GDP":3693.2,"CONT":"Europe","IND_DAY":null},"FRA":{"COUNTRY":"France","POP":67.02,"AREA":640.68,"GDP":2582.49,"CONT":"Europe","IND_DAY":"1789-07-14"},"GBR":{"COUNTRY":"UK","POP":66.44,"AREA":242.5,"GDP":2631.23,"CONT":"Europe","IND_DAY":null},"ITA":{"COUNTRY":"Italy","POP":60.36,"AREA":301.34,"GDP":1943.84,"CONT":"Europe","IND_DAY":null},"ARG":{"COUNTRY":"Argentina","POP":44.94,"AREA":2780.4,"GDP":637.49,"CONT":"S.America","IND_DAY":"1816-07-09"},"DZA":{"COUNTRY":"Algeria","POP":43.38,"AREA":2381.74,"GDP":167.56,"CONT":"Africa","IND_DAY":"1962-07-05"},"CAN":{"COUNTRY":"Canada","POP":37.59,"AREA":9984.67,"GDP":1647.12,"CONT":"N.America","IND_DAY":"1867-07-01"},"AUS":{"COUNTRY":"Australia","POP":25.47,"AREA":7692.02,"GDP":1408.68,"CONT":"Oceania","IND_DAY":null},"KAZ":{"COUNTRY":"Kazakhstan","POP":18.53,"AREA":2724.9,"GDP":159.41,"CONT":"Asia","IND_DAY":"1991-12-16"}}
data-index.json also has one large dictionary, but this time the row labels are the keys, and the inner dictionaries are the values.
There are few more options for orient. One of them is 'records':
>>>
>>> df.to_json('data-records.json', orient='records')
This code should yield the file data-records.json. You can expand the code block below to see the content:
[{"COUNTRY":"China","POP":1398.72,"AREA":9596.96,"GDP":12234.78,"CONT":"Asia","IND_DAY":null},{"COUNTRY":"India","POP":1351.16,"AREA":3287.26,"GDP":2575.67,"CONT":"Asia","IND_DAY":"1947-08-15"},{"COUNTRY":"US","POP":329.74,"AREA":9833.52,"GDP":19485.39,"CONT":"N.America","IND_DAY":"1776-07-04"},{"COUNTRY":"Indonesia","POP":268.07,"AREA":1910.93,"GDP":1015.54,"CONT":"Asia","IND_DAY":"1945-08-17"},{"COUNTRY":"Brazil","POP":210.32,"AREA":8515.77,"GDP":2055.51,"CONT":"S.America","IND_DAY":"1822-09-07"},{"COUNTRY":"Pakistan","POP":205.71,"AREA":881.91,"GDP":302.14,"CONT":"Asia","IND_DAY":"1947-08-14"},{"COUNTRY":"Nigeria","POP":200.96,"AREA":923.77,"GDP":375.77,"CONT":"Africa","IND_DAY":"1960-10-01"},{"COUNTRY":"Bangladesh","POP":167.09,"AREA":147.57,"GDP":245.63,"CONT":"Asia","IND_DAY":"1971-03-26"},{"COUNTRY":"Russia","POP":146.79,"AREA":17098.25,"GDP":1530.75,"CONT":null,"IND_DAY":"1992-06-12"},{"COUNTRY":"Mexico","POP":126.58,"AREA":1964.38,"GDP":1158.23,"CONT":"N.America","IND_DAY":"1810-09-16"},{"COUNTRY":"Japan","POP":126.22,"AREA":377.97,"GDP":4872.42,"CONT":"Asia","IND_DAY":null},{"COUNTRY":"Germany","POP":83.02,"AREA":357.11,"GDP":3693.2,"CONT":"Europe","IND_DAY":null},{"COUNTRY":"France","POP":67.02,"AREA":640.68,"GDP":2582.49,"CONT":"Europe","IND_DAY":"1789-07-14"},{"COUNTRY":"UK","POP":66.44,"AREA":242.5,"GDP":2631.23,"CONT":"Europe","IND_DAY":null},{"COUNTRY":"Italy","POP":60.36,"AREA":301.34,"GDP":1943.84,"CONT":"Europe","IND_DAY":null},{"COUNTRY":"Argentina","POP":44.94,"AREA":2780.4,"GDP":637.49,"CONT":"S.America","IND_DAY":"1816-07-09"},{"COUNTRY":"Algeria","POP":43.38,"AREA":2381.74,"GDP":167.56,"CONT":"Africa","IND_DAY":"1962-07-05"},{"COUNTRY":"Canada","POP":37.59,"AREA":9984.67,"GDP":1647.12,"CONT":"N.America","IND_DAY":"1867-07-01"},{"COUNTRY":"Australia","POP":25.47,"AREA":7692.02,"GDP":1408.68,"CONT":"Oceania","IND_DAY":null},{"COUNTRY":"Kazakhstan","POP":18.53,"AREA":2724.9,"GDP":159.41,"CONT":"Asia","IND_DAY":"1991-12-16"}]
data-records.json holds a list with one dictionary for each row. The row labels are not written.
You can get another interesting file structure with orient='split':
>>>
>>> df.to_json('data-split.json', orient='split')
The resulting file is data-split.json. You can expand the code block below to see how this file should look:
{"columns":["COUNTRY","POP","AREA","GDP","CONT","IND_DAY"],"index":["CHN","IND","USA","IDN","BRA","PAK","NGA","BGD","RUS","MEX","JPN","DEU","FRA","GBR","ITA","ARG","DZA","CAN","AUS","KAZ"],"data":[["China",1398.72,9596.96,12234.78,"Asia",null],["India",1351.16,3287.26,2575.67,"Asia","1947-08-15"],["US",329.74,9833.52,19485.39,"N.America","1776-07-04"],["Indonesia",268.07,1910.93,1015.54,"Asia","1945-08-17"],["Brazil",210.32,8515.77,2055.51,"S.America","1822-09-07"],["Pakistan",205.71,881.91,302.14,"Asia","1947-08-14"],["Nigeria",200.96,923.77,375.77,"Africa","1960-10-01"],["Bangladesh",167.09,147.57,245.63,"Asia","1971-03-26"],["Russia",146.79,17098.25,1530.75,null,"1992-06-12"],["Mexico",126.58,1964.38,1158.23,"N.America","1810-09-16"],["Japan",126.22,377.97,4872.42,"Asia",null],["Germany",83.02,357.11,3693.2,"Europe",null],["France",67.02,640.68,2582.49,"Europe","1789-07-14"],["UK",66.44,242.5,2631.23,"Europe",null],["Italy",60.36,301.34,1943.84,"Europe",null],["Argentina",44.94,2780.4,637.49,"S.America","1816-07-09"],["Algeria",43.38,2381.74,167.56,"Africa","1962-07-05"],["Canada",37.59,9984.67,1647.12,"N.America","1867-07-01"],["Australia",25.47,7692.02,1408.68,"Oceania",null],["Kazakhstan",18.53,2724.9,159.41,"Asia","1991-12-16"]]}
data-split.json contains one dictionary that holds the following lists:
- The names of the columns
- The labels of the rows
- The inner lists (two-dimensional sequence) that hold data values
If you don’t provide the value for the optional parameter path_or_buf that defines the file path, then .to_json() will return a JSON string instead of writing the results to a file. This behavior is consistent with .to_csv().
There are other optional parameters you can use. For instance, you can set index=False to forgo saving row labels. You can manipulate precision with double_precision, and dates with date_format and date_unit. These last two parameters are particularly important when you have time series among your data:
>>>
>>> df = pd.DataFrame(data=data).T
>>> df['IND_DAY'] = pd.to_datetime(df['IND_DAY'])
>>> df.dtypes
COUNTRY object
POP object
AREA object
GDP object
CONT object
IND_DAY datetime64[ns]
dtype: object
>>> df.to_json('data-time.json')
In this example, you’ve created the DataFrame from the dictionary data and used to_datetime() to convert the values in the last column to datetime64. You can expand the code block below to see the resulting file:
{"COUNTRY":{"CHN":"China","IND":"India","USA":"US","IDN":"Indonesia","BRA":"Brazil","PAK":"Pakistan","NGA":"Nigeria","BGD":"Bangladesh","RUS":"Russia","MEX":"Mexico","JPN":"Japan","DEU":"Germany","FRA":"France","GBR":"UK","ITA":"Italy","ARG":"Argentina","DZA":"Algeria","CAN":"Canada","AUS":"Australia","KAZ":"Kazakhstan"},"POP":{"CHN":1398.72,"IND":1351.16,"USA":329.74,"IDN":268.07,"BRA":210.32,"PAK":205.71,"NGA":200.96,"BGD":167.09,"RUS":146.79,"MEX":126.58,"JPN":126.22,"DEU":83.02,"FRA":67.02,"GBR":66.44,"ITA":60.36,"ARG":44.94,"DZA":43.38,"CAN":37.59,"AUS":25.47,"KAZ":18.53},"AREA":{"CHN":9596.96,"IND":3287.26,"USA":9833.52,"IDN":1910.93,"BRA":8515.77,"PAK":881.91,"NGA":923.77,"BGD":147.57,"RUS":17098.25,"MEX":1964.38,"JPN":377.97,"DEU":357.11,"FRA":640.68,"GBR":242.5,"ITA":301.34,"ARG":2780.4,"DZA":2381.74,"CAN":9984.67,"AUS":7692.02,"KAZ":2724.9},"GDP":{"CHN":12234.78,"IND":2575.67,"USA":19485.39,"IDN":1015.54,"BRA":2055.51,"PAK":302.14,"NGA":375.77,"BGD":245.63,"RUS":1530.75,"MEX":1158.23,"JPN":4872.42,"DEU":3693.2,"FRA":2582.49,"GBR":2631.23,"ITA":1943.84,"ARG":637.49,"DZA":167.56,"CAN":1647.12,"AUS":1408.68,"KAZ":159.41},"CONT":{"CHN":"Asia","IND":"Asia","USA":"N.America","IDN":"Asia","BRA":"S.America","PAK":"Asia","NGA":"Africa","BGD":"Asia","RUS":null,"MEX":"N.America","JPN":"Asia","DEU":"Europe","FRA":"Europe","GBR":"Europe","ITA":"Europe","ARG":"S.America","DZA":"Africa","CAN":"N.America","AUS":"Oceania","KAZ":"Asia"},"IND_DAY":{"CHN":null,"IND":-706320000000,"USA":-6106060800000,"IDN":-769219200000,"BRA":-4648924800000,"PAK":-706406400000,"NGA":-291945600000,"BGD":38793600000,"RUS":708307200000,"MEX":-5026838400000,"JPN":null,"DEU":null,"FRA":-5694969600000,"GBR":null,"ITA":null,"ARG":-4843411200000,"DZA":-236476800000,"CAN":-3234729600000,"AUS":null,"KAZ":692841600000}}
In this file, you have large integers instead of dates for the independence days. That’s because the default value of the optional parameter date_format is 'epoch' whenever orient isn’t 'table'. This default behavior expresses dates as an epoch in milliseconds relative to midnight on January 1, 1970.
However, if you pass date_format='iso', then you’ll get the dates in the ISO 8601 format. In addition, date_unit decides the units of time:
>>>
>>> df = pd.DataFrame(data=data).T
>>> df['IND_DAY'] = pd.to_datetime(df['IND_DAY'])
>>> df.to_json('new-data-time.json', date_format='iso', date_unit='s')
This code produces the following JSON file:
{"COUNTRY":{"CHN":"China","IND":"India","USA":"US","IDN":"Indonesia","BRA":"Brazil","PAK":"Pakistan","NGA":"Nigeria","BGD":"Bangladesh","RUS":"Russia","MEX":"Mexico","JPN":"Japan","DEU":"Germany","FRA":"France","GBR":"UK","ITA":"Italy","ARG":"Argentina","DZA":"Algeria","CAN":"Canada","AUS":"Australia","KAZ":"Kazakhstan"},"POP":{"CHN":1398.72,"IND":1351.16,"USA":329.74,"IDN":268.07,"BRA":210.32,"PAK":205.71,"NGA":200.96,"BGD":167.09,"RUS":146.79,"MEX":126.58,"JPN":126.22,"DEU":83.02,"FRA":67.02,"GBR":66.44,"ITA":60.36,"ARG":44.94,"DZA":43.38,"CAN":37.59,"AUS":25.47,"KAZ":18.53},"AREA":{"CHN":9596.96,"IND":3287.26,"USA":9833.52,"IDN":1910.93,"BRA":8515.77,"PAK":881.91,"NGA":923.77,"BGD":147.57,"RUS":17098.25,"MEX":1964.38,"JPN":377.97,"DEU":357.11,"FRA":640.68,"GBR":242.5,"ITA":301.34,"ARG":2780.4,"DZA":2381.74,"CAN":9984.67,"AUS":7692.02,"KAZ":2724.9},"GDP":{"CHN":12234.78,"IND":2575.67,"USA":19485.39,"IDN":1015.54,"BRA":2055.51,"PAK":302.14,"NGA":375.77,"BGD":245.63,"RUS":1530.75,"MEX":1158.23,"JPN":4872.42,"DEU":3693.2,"FRA":2582.49,"GBR":2631.23,"ITA":1943.84,"ARG":637.49,"DZA":167.56,"CAN":1647.12,"AUS":1408.68,"KAZ":159.41},"CONT":{"CHN":"Asia","IND":"Asia","USA":"N.America","IDN":"Asia","BRA":"S.America","PAK":"Asia","NGA":"Africa","BGD":"Asia","RUS":null,"MEX":"N.America","JPN":"Asia","DEU":"Europe","FRA":"Europe","GBR":"Europe","ITA":"Europe","ARG":"S.America","DZA":"Africa","CAN":"N.America","AUS":"Oceania","KAZ":"Asia"},"IND_DAY":{"CHN":null,"IND":"1947-08-15T00:00:00Z","USA":"1776-07-04T00:00:00Z","IDN":"1945-08-17T00:00:00Z","BRA":"1822-09-07T00:00:00Z","PAK":"1947-08-14T00:00:00Z","NGA":"1960-10-01T00:00:00Z","BGD":"1971-03-26T00:00:00Z","RUS":"1992-06-12T00:00:00Z","MEX":"1810-09-16T00:00:00Z","JPN":null,"DEU":null,"FRA":"1789-07-14T00:00:00Z","GBR":null,"ITA":null,"ARG":"1816-07-09T00:00:00Z","DZA":"1962-07-05T00:00:00Z","CAN":"1867-07-01T00:00:00Z","AUS":null,"KAZ":"1991-12-16T00:00:00Z"}}
The dates in the resulting file are in the ISO 8601 format.
You can load the data from a JSON file with read_json():
>>>
>>> df = pd.read_json('data-index.json', orient='index',
... convert_dates=['IND_DAY'])
The parameter convert_dates has a similar purpose as parse_dates when you use it to read CSV files. The optional parameter orient is very important because it specifies how pandas understands the structure of the file.
There are other optional parameters you can use as well:
- Set the encoding with
encoding. - Manipulate dates with
convert_datesandkeep_default_dates. - Impact precision with
dtypeandprecise_float. - Decode numeric data directly to NumPy arrays with
numpy=True.
Note that you might lose the order of rows and columns when using the JSON format to store your data.
HTML Files
An HTML is a plaintext file that uses hypertext markup language to help browsers render web pages. The extensions for HTML files are .html and .htm. You’ll need to install an HTML parser library like lxml or html5lib to be able to work with HTML files:
$pip install lxml html5lib
You can also use Conda to install the same packages:
$ conda install lxml html5lib
Once you have these libraries, you can save the contents of your DataFrame as an HTML file with .to_html():
>>>
df = pd.DataFrame(data=data).T
df.to_html('data.html')
This code generates a file data.html. You can expand the code block below to see how this file should look:
<table border="1" class="dataframe">
<thead>
<tr style="text-align: right;">
<th></th>
<th>COUNTRY</th>
<th>POP</th>
<th>AREA</th>
<th>GDP</th>
<th>CONT</th>
<th>IND_DAY</th>
</tr>
</thead>
<tbody>
<tr>
<th>CHN</th>
<td>China</td>
<td>1398.72</td>
<td>9596.96</td>
<td>12234.8</td>
<td>Asia</td>
<td>NaN</td>
</tr>
<tr>
<th>IND</th>
<td>India</td>
<td>1351.16</td>
<td>3287.26</td>
<td>2575.67</td>
<td>Asia</td>
<td>1947-08-15</td>
</tr>
<tr>
<th>USA</th>
<td>US</td>
<td>329.74</td>
<td>9833.52</td>
<td>19485.4</td>
<td>N.America</td>
<td>1776-07-04</td>
</tr>
<tr>
<th>IDN</th>
<td>Indonesia</td>
<td>268.07</td>
<td>1910.93</td>
<td>1015.54</td>
<td>Asia</td>
<td>1945-08-17</td>
</tr>
<tr>
<th>BRA</th>
<td>Brazil</td>
<td>210.32</td>
<td>8515.77</td>
<td>2055.51</td>
<td>S.America</td>
<td>1822-09-07</td>
</tr>
<tr>
<th>PAK</th>
<td>Pakistan</td>
<td>205.71</td>
<td>881.91</td>
<td>302.14</td>
<td>Asia</td>
<td>1947-08-14</td>
</tr>
<tr>
<th>NGA</th>
<td>Nigeria</td>
<td>200.96</td>
<td>923.77</td>
<td>375.77</td>
<td>Africa</td>
<td>1960-10-01</td>
</tr>
<tr>
<th>BGD</th>
<td>Bangladesh</td>
<td>167.09</td>
<td>147.57</td>
<td>245.63</td>
<td>Asia</td>
<td>1971-03-26</td>
</tr>
<tr>
<th>RUS</th>
<td>Russia</td>
<td>146.79</td>
<td>17098.2</td>
<td>1530.75</td>
<td>NaN</td>
<td>1992-06-12</td>
</tr>
<tr>
<th>MEX</th>
<td>Mexico</td>
<td>126.58</td>
<td>1964.38</td>
<td>1158.23</td>
<td>N.America</td>
<td>1810-09-16</td>
</tr>
<tr>
<th>JPN</th>
<td>Japan</td>
<td>126.22</td>
<td>377.97</td>
<td>4872.42</td>
<td>Asia</td>
<td>NaN</td>
</tr>
<tr>
<th>DEU</th>
<td>Germany</td>
<td>83.02</td>
<td>357.11</td>
<td>3693.2</td>
<td>Europe</td>
<td>NaN</td>
</tr>
<tr>
<th>FRA</th>
<td>France</td>
<td>67.02</td>
<td>640.68</td>
<td>2582.49</td>
<td>Europe</td>
<td>1789-07-14</td>
</tr>
<tr>
<th>GBR</th>
<td>UK</td>
<td>66.44</td>
<td>242.5</td>
<td>2631.23</td>
<td>Europe</td>
<td>NaN</td>
</tr>
<tr>
<th>ITA</th>
<td>Italy</td>
<td>60.36</td>
<td>301.34</td>
<td>1943.84</td>
<td>Europe</td>
<td>NaN</td>
</tr>
<tr>
<th>ARG</th>
<td>Argentina</td>
<td>44.94</td>
<td>2780.4</td>
<td>637.49</td>
<td>S.America</td>
<td>1816-07-09</td>
</tr>
<tr>
<th>DZA</th>
<td>Algeria</td>
<td>43.38</td>
<td>2381.74</td>
<td>167.56</td>
<td>Africa</td>
<td>1962-07-05</td>
</tr>
<tr>
<th>CAN</th>
<td>Canada</td>
<td>37.59</td>
<td>9984.67</td>
<td>1647.12</td>
<td>N.America</td>
<td>1867-07-01</td>
</tr>
<tr>
<th>AUS</th>
<td>Australia</td>
<td>25.47</td>
<td>7692.02</td>
<td>1408.68</td>
<td>Oceania</td>
<td>NaN</td>
</tr>
<tr>
<th>KAZ</th>
<td>Kazakhstan</td>
<td>18.53</td>
<td>2724.9</td>
<td>159.41</td>
<td>Asia</td>
<td>1991-12-16</td>
</tr>
</tbody>
</table>
This file shows the DataFrame contents nicely. However, notice that you haven’t obtained an entire web page. You’ve just output the data that corresponds to df in the HTML format.
.to_html() won’t create a file if you don’t provide the optional parameter buf, which denotes the buffer to write to. If you leave this parameter out, then your code will return a string as it did with .to_csv() and .to_json().
Here are some other optional parameters:
headerdetermines whether to save the column names.indexdetermines whether to save the row labels.classesassigns cascading style sheet (CSS) classes.render_linksspecifies whether to convert URLs to HTML links.table_idassigns the CSSidto thetabletag.escapedecides whether to convert the characters<,>, and&to HTML-safe strings.
You use parameters like these to specify different aspects of the resulting files or strings.
You can create a DataFrame object from a suitable HTML file using read_html(), which will return a DataFrame instance or a list of them:
>>>
>>> df = pd.read_html('data.html', index_col=0, parse_dates=['IND_DAY'])
This is very similar to what you did when reading CSV files. You also have parameters that help you work with dates, missing values, precision, encoding, HTML parsers, and more.
Excel Files
You’ve already learned how to read and write Excel files with pandas. However, there are a few more options worth considering. For one, when you use .to_excel(), you can specify the name of the target worksheet with the optional parameter sheet_name:
>>>
>>> df = pd.DataFrame(data=data).T
>>> df.to_excel('data.xlsx', sheet_name='COUNTRIES')
Here, you create a file data.xlsx with a worksheet called COUNTRIES that stores the data. The string 'data.xlsx' is the argument for the parameter excel_writer that defines the name of the Excel file or its path.
The optional parameters startrow and startcol both default to 0 and indicate the upper left-most cell where the data should start being written:
>>>
>>> df.to_excel('data-shifted.xlsx', sheet_name='COUNTRIES',
... startrow=2, startcol=4)
Here, you specify that the table should start in the third row and the fifth column. You also used zero-based indexing, so the third row is denoted by 2 and the fifth column by 4.
Now the resulting worksheet looks like this:
As you can see, the table starts in the third row 2 and the fifth column E.
.read_excel() also has the optional parameter sheet_name that specifies which worksheets to read when loading data. It can take on one of the following values:
- The zero-based index of the worksheet
- The name of the worksheet
- The list of indices or names to read multiple sheets
- The value
Noneto read all sheets
Here’s how you would use this parameter in your code:
>>>
>>> df = pd.read_excel('data.xlsx', sheet_name=0, index_col=0,
... parse_dates=['IND_DAY'])
>>> df = pd.read_excel('data.xlsx', sheet_name='COUNTRIES', index_col=0,
... parse_dates=['IND_DAY'])
Both statements above create the same DataFrame because the sheet_name parameters have the same values. In both cases, sheet_name=0 and sheet_name='COUNTRIES' refer to the same worksheet. The argument parse_dates=['IND_DAY'] tells pandas to try to consider the values in this column as dates or times.
There are other optional parameters you can use with .read_excel() and .to_excel() to determine the Excel engine, the encoding, the way to handle missing values and infinities, the method for writing column names and row labels, and so on.
SQL Files
pandas IO tools can also read and write databases. In this next example, you’ll write your data to a database called data.db. To get started, you’ll need the SQLAlchemy package. To learn more about it, you can read the official ORM tutorial. You’ll also need the database driver. Python has a built-in driver for SQLite.
You can install SQLAlchemy with pip:
You can also install it with Conda:
$ conda install sqlalchemy
Once you have SQLAlchemy installed, import create_engine() and create a database engine:
>>>
>>> from sqlalchemy import create_engine
>>> engine = create_engine('sqlite:///data.db', echo=False)
Now that you have everything set up, the next step is to create a DataFrame object. It’s convenient to specify the data types and apply .to_sql().
>>>
>>> dtypes = {'POP': 'float64', 'AREA': 'float64', 'GDP': 'float64',
... 'IND_DAY': 'datetime64'}
>>> df = pd.DataFrame(data=data).T.astype(dtype=dtypes)
>>> df.dtypes
COUNTRY object
POP float64
AREA float64
GDP float64
CONT object
IND_DAY datetime64[ns]
dtype: object
.astype() is a very convenient method you can use to set multiple data types at once.
Once you’ve created your DataFrame, you can save it to the database with .to_sql():
>>>
>>> df.to_sql('data.db', con=engine, index_label='ID')
The parameter con is used to specify the database connection or engine that you want to use. The optional parameter index_label specifies how to call the database column with the row labels. You’ll often see it take on the value ID, Id, or id.
You should get the database data.db with a single table that looks like this:
The first column contains the row labels. To omit writing them into the database, pass index=False to .to_sql(). The other columns correspond to the columns of the DataFrame.
There are a few more optional parameters. For example, you can use schema to specify the database schema and dtype to determine the types of the database columns. You can also use if_exists, which says what to do if a database with the same name and path already exists:
if_exists='fail'raises a ValueError and is the default.if_exists='replace'drops the table and inserts new values.if_exists='append'inserts new values into the table.
You can load the data from the database with read_sql():
>>>
>>> df = pd.read_sql('data.db', con=engine, index_col='ID')
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
ID
CHN China 1398.72 9596.96 12234.78 Asia NaT
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.25 1530.75 None 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaT
DEU Germany 83.02 357.11 3693.20 Europe NaT
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaT
ITA Italy 60.36 301.34 1943.84 Europe NaT
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaT
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
The parameter index_col specifies the name of the column with the row labels. Note that this inserts an extra row after the header that starts with ID. You can fix this behavior with the following line of code:
>>>
>>> df.index.name = None
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.78 Asia NaT
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.25 1530.75 None 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaT
DEU Germany 83.02 357.11 3693.20 Europe NaT
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaT
ITA Italy 60.36 301.34 1943.84 Europe NaT
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaT
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
Now you have the same DataFrame object as before.
Note that the continent for Russia is now None instead of nan. If you want to fill the missing values with nan, then you can use .fillna():
>>>
>>> df.fillna(value=float('nan'), inplace=True)
.fillna() replaces all missing values with whatever you pass to value. Here, you passed float('nan'), which says to fill all missing values with nan.
Also note that you didn’t have to pass parse_dates=['IND_DAY'] to read_sql(). That’s because your database was able to detect that the last column contains dates. However, you can pass parse_dates if you’d like. You’ll get the same results.
There are other functions that you can use to read databases, like read_sql_table() and read_sql_query(). Feel free to try them out!
Pickle Files
Pickling is the act of converting Python objects into byte streams. Unpickling is the inverse process. Python pickle files are the binary files that keep the data and hierarchy of Python objects. They usually have the extension .pickle or .pkl.
You can save your DataFrame in a pickle file with .to_pickle():
>>>
>>> dtypes = {'POP': 'float64', 'AREA': 'float64', 'GDP': 'float64',
... 'IND_DAY': 'datetime64'}
>>> df = pd.DataFrame(data=data).T.astype(dtype=dtypes)
>>> df.to_pickle('data.pickle')
Like you did with databases, it can be convenient first to specify the data types. Then, you create a file data.pickle to contain your data. You could also pass an integer value to the optional parameter protocol, which specifies the protocol of the pickler.
You can get the data from a pickle file with read_pickle():
>>>
>>> df = pd.read_pickle('data.pickle')
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.78 Asia NaT
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaT
DEU Germany 83.02 357.11 3693.20 Europe NaT
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaT
ITA Italy 60.36 301.34 1943.84 Europe NaT
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaT
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
read_pickle() returns the DataFrame with the stored data. You can also check the data types:
>>>
>>> df.dtypes
COUNTRY object
POP float64
AREA float64
GDP float64
CONT object
IND_DAY datetime64[ns]
dtype: object
These are the same ones that you specified before using .to_pickle().
As a word of caution, you should always beware of loading pickles from untrusted sources. This can be dangerous! When you unpickle an untrustworthy file, it could execute arbitrary code on your machine, gain remote access to your computer, or otherwise exploit your device in other ways.
Working With Big Data
If your files are too large for saving or processing, then there are several approaches you can take to reduce the required disk space:
- Compress your files
- Choose only the columns you want
- Omit the rows you don’t need
- Force the use of less precise data types
- Split the data into chunks
You’ll take a look at each of these techniques in turn.
Compress and Decompress Files
You can create an archive file like you would a regular one, with the addition of a suffix that corresponds to the desired compression type:
'.gz''.bz2''.zip''.xz'
pandas can deduce the compression type by itself:
>>>
>>> df = pd.DataFrame(data=data).T
>>> df.to_csv('data.csv.zip')
Here, you create a compressed .csv file as an archive. The size of the regular .csv file is 1048 bytes, while the compressed file only has 766 bytes.
You can open this compressed file as usual with the pandas read_csv() function:
>>>
>>> df = pd.read_csv('data.csv.zip', index_col=0,
... parse_dates=['IND_DAY'])
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.78 Asia NaT
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaT
DEU Germany 83.02 357.11 3693.20 Europe NaT
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaT
ITA Italy 60.36 301.34 1943.84 Europe NaT
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaT
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
read_csv() decompresses the file before reading it into a DataFrame.
You can specify the type of compression with the optional parameter compression, which can take on any of the following values:
'infer''gzip''bz2''zip''xz'None
The default value compression='infer' indicates that pandas should deduce the compression type from the file extension.
Here’s how you would compress a pickle file:
>>>
>>> df = pd.DataFrame(data=data).T
>>> df.to_pickle('data.pickle.compress', compression='gzip')
You should get the file data.pickle.compress that you can later decompress and read:
>>>
>>> df = pd.read_pickle('data.pickle.compress', compression='gzip')
df again corresponds to the DataFrame with the same data as before.
You can give the other compression methods a try, as well. If you’re using pickle files, then keep in mind that the .zip format supports reading only.
Choose Columns
The pandas read_csv() and read_excel() functions have the optional parameter usecols that you can use to specify the columns you want to load from the file. You can pass the list of column names as the corresponding argument:
>>>
>>> df = pd.read_csv('data.csv', usecols=['COUNTRY', 'AREA'])
>>> df
COUNTRY AREA
0 China 9596.96
1 India 3287.26
2 US 9833.52
3 Indonesia 1910.93
4 Brazil 8515.77
5 Pakistan 881.91
6 Nigeria 923.77
7 Bangladesh 147.57
8 Russia 17098.25
9 Mexico 1964.38
10 Japan 377.97
11 Germany 357.11
12 France 640.68
13 UK 242.50
14 Italy 301.34
15 Argentina 2780.40
16 Algeria 2381.74
17 Canada 9984.67
18 Australia 7692.02
19 Kazakhstan 2724.90
Now you have a DataFrame that contains less data than before. Here, there are only the names of the countries and their areas.
Instead of the column names, you can also pass their indices:
>>>
>>> df = pd.read_csv('data.csv',index_col=0, usecols=[0, 1, 3])
>>> df
COUNTRY AREA
CHN China 9596.96
IND India 3287.26
USA US 9833.52
IDN Indonesia 1910.93
BRA Brazil 8515.77
PAK Pakistan 881.91
NGA Nigeria 923.77
BGD Bangladesh 147.57
RUS Russia 17098.25
MEX Mexico 1964.38
JPN Japan 377.97
DEU Germany 357.11
FRA France 640.68
GBR UK 242.50
ITA Italy 301.34
ARG Argentina 2780.40
DZA Algeria 2381.74
CAN Canada 9984.67
AUS Australia 7692.02
KAZ Kazakhstan 2724.90
Expand the code block below to compare these results with the file 'data.csv':
,COUNTRY,POP,AREA,GDP,CONT,IND_DAY
CHN,China,1398.72,9596.96,12234.78,Asia,
IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15
USA,US,329.74,9833.52,19485.39,N.America,1776-07-04
IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17
BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07
PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14
NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01
BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26
RUS,Russia,146.79,17098.25,1530.75,,1992-06-12
MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16
JPN,Japan,126.22,377.97,4872.42,Asia,
DEU,Germany,83.02,357.11,3693.2,Europe,
FRA,France,67.02,640.68,2582.49,Europe,1789-07-14
GBR,UK,66.44,242.5,2631.23,Europe,
ITA,Italy,60.36,301.34,1943.84,Europe,
ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09
DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05
CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01
AUS,Australia,25.47,7692.02,1408.68,Oceania,
KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
You can see the following columns:
- The column at index
0contains the row labels. - The column at index
1contains the country names. - The column at index
3contains the areas.
Simlarly, read_sql() has the optional parameter columns that takes a list of column names to read:
>>>
>>> df = pd.read_sql('data.db', con=engine, index_col='ID',
... columns=['COUNTRY', 'AREA'])
>>> df.index.name = None
>>> df
COUNTRY AREA
CHN China 9596.96
IND India 3287.26
USA US 9833.52
IDN Indonesia 1910.93
BRA Brazil 8515.77
PAK Pakistan 881.91
NGA Nigeria 923.77
BGD Bangladesh 147.57
RUS Russia 17098.25
MEX Mexico 1964.38
JPN Japan 377.97
DEU Germany 357.11
FRA France 640.68
GBR UK 242.50
ITA Italy 301.34
ARG Argentina 2780.40
DZA Algeria 2381.74
CAN Canada 9984.67
AUS Australia 7692.02
KAZ Kazakhstan 2724.90
Again, the DataFrame only contains the columns with the names of the countries and areas. If columns is None or omitted, then all of the columns will be read, as you saw before. The default behavior is columns=None.
Omit Rows
When you test an algorithm for data processing or machine learning, you often don’t need the entire dataset. It’s convenient to load only a subset of the data to speed up the process. The pandas read_csv() and read_excel() functions have some optional parameters that allow you to select which rows you want to load:
skiprows: either the number of rows to skip at the beginning of the file if it’s an integer, or the zero-based indices of the rows to skip if it’s a list-like objectskipfooter: the number of rows to skip at the end of the filenrows: the number of rows to read
Here’s how you would skip rows with odd zero-based indices, keeping the even ones:
>>>
>>> df = pd.read_csv('data.csv', index_col=0, skiprows=range(1, 20, 2))
>>> df
COUNTRY POP AREA GDP CONT IND_DAY
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
DEU Germany 83.02 357.11 3693.20 Europe NaN
GBR UK 66.44 242.50 2631.23 Europe NaN
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
In this example, skiprows is range(1, 20, 2) and corresponds to the values 1, 3, …, 19. The instances of the Python built-in class range behave like sequences. The first row of the file data.csv is the header row. It has the index 0, so pandas loads it in. The second row with index 1 corresponds to the label CHN, and pandas skips it. The third row with the index 2 and label IND is loaded, and so on.
If you want to choose rows randomly, then skiprows can be a list or NumPy array with pseudo-random numbers, obtained either with pure Python or with NumPy.
Force Less Precise Data Types
If you’re okay with less precise data types, then you can potentially save a significant amount of memory! First, get the data types with .dtypes again:
>>>
>>> df = pd.read_csv('data.csv', index_col=0, parse_dates=['IND_DAY'])
>>> df.dtypes
COUNTRY object
POP float64
AREA float64
GDP float64
CONT object
IND_DAY datetime64[ns]
dtype: object
The columns with the floating-point numbers are 64-bit floats. Each number of this type float64 consumes 64 bits or 8 bytes. Each column has 20 numbers and requires 160 bytes. You can verify this with .memory_usage():
>>>
>>> df.memory_usage()
Index 160
COUNTRY 160
POP 160
AREA 160
GDP 160
CONT 160
IND_DAY 160
dtype: int64
.memory_usage() returns an instance of Series with the memory usage of each column in bytes. You can conveniently combine it with .loc[] and .sum() to get the memory for a group of columns:
>>>
>>> df.loc[:, ['POP', 'AREA', 'GDP']].memory_usage(index=False).sum()
480
This example shows how you can combine the numeric columns 'POP', 'AREA', and 'GDP' to get their total memory requirement. The argument index=False excludes data for row labels from the resulting Series object. For these three columns, you’ll need 480 bytes.
You can also extract the data values in the form of a NumPy array with .to_numpy() or .values. Then, use the .nbytes attribute to get the total bytes consumed by the items of the array:
>>>
>>> df.loc[:, ['POP', 'AREA', 'GDP']].to_numpy().nbytes
480
The result is the same 480 bytes. So, how do you save memory?
In this case, you can specify that your numeric columns 'POP', 'AREA', and 'GDP' should have the type float32. Use the optional parameter dtype to do this:
>>>
>>> dtypes = {'POP': 'float32', 'AREA': 'float32', 'GDP': 'float32'}
>>> df = pd.read_csv('data.csv', index_col=0, dtype=dtypes,
... parse_dates=['IND_DAY'])
The dictionary dtypes specifies the desired data types for each column. It’s passed to the pandas read_csv() function as the argument that corresponds to the parameter dtype.
Now you can verify that each numeric column needs 80 bytes, or 4 bytes per item:
>>>
>>> df.dtypes
COUNTRY object
POP float32
AREA float32
GDP float32
CONT object
IND_DAY datetime64[ns]
dtype: object
>>> df.memory_usage()
Index 160
COUNTRY 160
POP 80
AREA 80
GDP 80
CONT 160
IND_DAY 160
dtype: int64
>>> df.loc[:, ['POP', 'AREA', 'GDP']].memory_usage(index=False).sum()
240
>>> df.loc[:, ['POP', 'AREA', 'GDP']].to_numpy().nbytes
240
Each value is a floating-point number of 32 bits or 4 bytes. The three numeric columns contain 20 items each. In total, you’ll need 240 bytes of memory when you work with the type float32. This is half the size of the 480 bytes you’d need to work with float64.
In addition to saving memory, you can significantly reduce the time required to process data by using float32 instead of float64 in some cases.
Use Chunks to Iterate Through Files
Another way to deal with very large datasets is to split the data into smaller chunks and process one chunk at a time. If you use read_csv(), read_json() or read_sql(), then you can specify the optional parameter chunksize:
>>>
>>> data_chunk = pd.read_csv('data.csv', index_col=0, chunksize=8)
>>> type(data_chunk)
<class 'pandas.io.parsers.TextFileReader'>
>>> hasattr(data_chunk, '__iter__')
True
>>> hasattr(data_chunk, '__next__')
True
chunksize defaults to None and can take on an integer value that indicates the number of items in a single chunk. When chunksize is an integer, read_csv() returns an iterable that you can use in a for loop to get and process only a fragment of the dataset in each iteration:
>>>
>>> for df_chunk in pd.read_csv('data.csv', index_col=0, chunksize=8):
... print(df_chunk, end='nn')
... print('memory:', df_chunk.memory_usage().sum(), 'bytes',
... end='nnn')
...
COUNTRY POP AREA GDP CONT IND_DAY
CHN China 1398.72 9596.96 12234.78 Asia NaN
IND India 1351.16 3287.26 2575.67 Asia 1947-08-15
USA US 329.74 9833.52 19485.39 N.America 1776-07-04
IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17
BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07
PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14
NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01
BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26
memory: 448 bytes
COUNTRY POP AREA GDP CONT IND_DAY
RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12
MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16
JPN Japan 126.22 377.97 4872.42 Asia NaN
DEU Germany 83.02 357.11 3693.20 Europe NaN
FRA France 67.02 640.68 2582.49 Europe 1789-07-14
GBR UK 66.44 242.50 2631.23 Europe NaN
ITA Italy 60.36 301.34 1943.84 Europe NaN
ARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09
memory: 448 bytes
COUNTRY POP AREA GDP CONT IND_DAY
DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05
CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01
AUS Australia 25.47 7692.02 1408.68 Oceania NaN
KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
memory: 224 bytes
In this example, the chunksize is 8. The first iteration of the for loop returns a DataFrame with the first eight rows of the dataset only. The second iteration returns another DataFrame with the next eight rows. The third and last iteration returns the remaining four rows.
In each iteration, you get and process the DataFrame with the number of rows equal to chunksize. It’s possible to have fewer rows than the value of chunksize in the last iteration. You can use this functionality to control the amount of memory required to process data and keep that amount reasonably small.
Conclusion
You now know how to save the data and labels from pandas DataFrame objects to different kinds of files. You also know how to load your data from files and create DataFrame objects.
You’ve used the pandas read_csv() and .to_csv() methods to read and write CSV files. You also used similar methods to read and write Excel, JSON, HTML, SQL, and pickle files. These functions are very convenient and widely used. They allow you to save or load your data in a single function or method call.
You’ve also learned how to save time, memory, and disk space when working with large data files:
- Compress or decompress files
- Choose the rows and columns you want to load
- Use less precise data types
- Split data into chunks and process them one by one
You’ve mastered a significant step in the machine learning and data science process! If you have any questions or comments, then please put them in the comments section below.
Watch Now This tutorial has a related video course created by the Real Python team. Watch it together with the written tutorial to deepen your understanding: Reading and Writing Files With Pandas
In this Pandas tutorial, we will learn how to work with Excel files (e.g., xls) in Python. It will provide an overview of how to use Pandas to load xlsx files and write spreadsheets to Excel.
In the first section, we will go through, with examples, how to use Pandas read_excel to; 1) read an Excel file, 2) read specific columns from a spreadsheet, 3) read multiple spreadsheets, and combine them to one dataframe. Furthermore, we are going to learn how to read many Excel files, and how to convert data according to specific data types (e.g., using Pandas dtypes).
When we have done this, we will continue by learning how to use Pandas to write Excel files; how to name the sheets and how to write to multiple sheets. Make sure to check out the newer post about reading xlsx files in Python with openpyxl, as well.
- Read the How to import Excel into R blog post if you need an overview on how to read xlsx files into R dataframes.
How to Install Pandas
Before we continue with this Pandas read and write Excel files tutorial there is something we need to do; installing Pandas (and Python, of course, if it’s not installed). We can install Pandas using Pip, given that we have Pip installed, that is. See here how to install pip.
# Linux Users
pip install pandas
# Windows Users
python pip install pandasCode language: Bash (bash)Note, if pip is telling us that there’s a newer version of pip, we may want to upgrade it. In a recent post, we cover how to upgrade pip to the latest version. Finally, before going on to the next section, you can use pip to install a certain version (i.e., older) of a packages usch as Pandas.
Installing Anaconda Scientific Python Distribution
Another great option is to consider is to install the Anaconda Python distribution. This is really an easy and fast way to get started with computer science. No need to worry about installing the packages you need to do computer science separately.
Both of the above methods are explained in this tutorial. Now, in a more recent blog post, we also cover how to install a Python package using pip, conda, and Anaconda. In that post, you will find more information about installing Python packages.
How to Read Excel Files to Pandas Dataframes:
Can Pandas read xlsx files? The short answer is, of course, “yes”. In this section, we are going to learn how to read Excel files and spreadsheets to Pandas dataframe objects. All examples in this Pandas Excel tutorial use local files. Note, that read_excel also can also load Excel files from a URL to a dataframe. As always when working with Pandas, we have to start by importing the module:
import pandas as pdCode language: Python (python)Now it’s time to learn how to use Pandas read_excel to read in data from an Excel file. The easiest way to use this method is to pass the file name as a string. If we don’t pass any other parameters, such as sheet name, it will read the first sheet in the index. In the first example, we are not going to use any parameters:
# Pandas read xlsx
df = pd.read_excel('MLBPlayerSalaries.xlsx')
df.head()Code language: Python (python)
Here, Pandas read_excel method read the data from the Excel file into a Pandas dataframe object. We then stored this dataframe into a variable called df.
When using read_excel Pandas will, by default, assign a numeric index or row label to the dataframe, and as usual, when int comes to Python, the index will start with zero. We may have a reason to leave the default index as it is.
For instance, if your data doesn’t have a column with unique values that can serve as a better index. In case there is a column that would serve as a better index, we can override the default behavior.
Setting the Index Column when Reading xls File
This is done by setting the index_col parameter to a column. It takes a numeric value for setting a single column as index or a list of numeric values for creating a multi-index. In the example below, we use the column ‘Player’ as indices. Note, these are not unique and it may, thus, not make sense to use these values as indices.
df = pd.read_excel('MLBPlayerSalaries.xlsx', sheet_names='MLBPlayerSalaries', index_col='Player')Code language: Python (python)Now, if one or two of your columns, for instance, are objects you use Pandas to_datetime to convert a column, properly.
Importing an Excel File to Pandas in Two Easy Steps:
Time needed: 1 minute.
Here’s a quick answer to the How do you import an Excel file into Python using Pandas? Importing an Excel file into a Pandas dataframe basically only requires two steps, given that we know the path, or URL, to the Excel file:
- Import Pandas
In the script type import pandas as pd
- Use Pandas read_excel method
Next step is to type df = pd.read_excel(FILE_PATH_OR_URL)
Remember to change FILE_PATH_OR_URL to the path or the URL of the Excel file.
Now that we know how easy it is to load an Excel file into a Pandas dataframe we are going to continue with learning more about the read_excel method.
Reading Specific Columns using Pandas read_excel
When using Pandas read_excel we will automatically get all columns from an Excel file. If we, for some reason, don’t want to parse all columns in the Excel file, we can use the parameter usecols. Let’s say we want to create a dataframe with the columns Player, Salary, and Position, only. We can do this by adding 1, 3, and 4 in a list:
cols = [1, 2, 3]
df = pd.read_excel('MLBPlayerSalaries.xlsx', sheet_names='MLBPlayerSalaries', usecols=cols)
df.head()Code language: Python (python)
According to the read_excel documentation, we should be able to put in a string. For instance, cols=’Player:Position’ should give us the same results as above.
Handling Missing Data using Pandas read_excel
If our data has missing values in some cells and these missing values are coded in some way, like “Missing” we can use the na_values parameter.
Pandas Read Excel Example with Missing Data
In the example below, we are using the parameter na_values and we are putting in a string (i.e., “Missing’):
df = pd.read_excel('MLBPlayerSalaries_MD.xlsx', na_values="Missing", sheet_names='MLBPlayerSalaries', usecols=cols)
df.head()Code language: Python (python)
In the read excel examples above we used a dataset that can be downloaded from this page.
- Read the post Data manipulation with Pandas for three methods on data manipulation of dataframes, including missing data.
- Learn easy methods to clean data using Pandas and Pyjanitor
How to Skip Rows when Reading an Excel File
Now we will learn how to skip rows when loading an Excel file using Pandas. For this read excel example, we will use data that can be downloaded here.
In the following Pandas read_excel example we load the sheet ‘session1’, which contains rows that we need to skip (these rows contain some information about the dataset).
We will use the parameter sheet_name=’Session1′ to read the sheet named ‘Session1’ (the example data contains more sheets; e.g., ‘Session2’ will load that sheet). Note, the first sheet will be read if we don’t use the sheet_name parameter. In this example, the important part is the parameter skiprow=2. We use this to skip the first two rows:
df = pd.read_excel('example_sheets1.xlsx', sheet_name='Session1', skiprows=2)
df.head()Code language: Python (python)
Another way to get Pandas read_excel to read from the Nth row is by using the header parameter. In the example Excel file, we use here, the third row contains the headers and we will use the parameter header=2 to tell Pandas read_excel that our headers are on the third row.
df = pd.read_excel('example_sheets1.xlsx', sheet_name='Session1', header=2)Code language: Python (python)Now, if we want Pandas read_excel to read from the second row, we change the number in the skiprows and header arguments to 2, and so on.
Reading Multiple Excel Sheets to Pandas Dataframes
In this section, of the Pandas read excel tutorial, we are going to learn how to read multiple sheets. Our Excel file, example_sheets1.xlsx’, has two sheets: ‘Session1’, and ‘Session2.’ Each sheet has data from an imagined experimental session. In the next example we are going to read both sheets, ‘Session1’ and ‘Session2’. Here’s how to use Pandas read_excel to read multiple sheets:
df = pd.read_excel('example_sheets1.xlsx',
sheet_name=['Session1', 'Session2'], skiprows=2)Code language: Python (python)By using the parameter sheet_name, and a list of names, we will get an ordered dictionary containing two dataframes:
dfCode language: Python (python)
When working with Pandas read_excel we may want to join the data from all sheets (in this case sessions). Merging Pandas dataframes are quite easy; we just use the concat function and loop over the keys (i.e., sheets):
df2 = pd.concat(df[frame] for frame in data.keys())Code language: Python (python)Now in the example Excel file, there is a column identifying the dataset (e.g., session number). However, maybe we don’t have that kind of information in our Excel file. To merge the two dataframes and adding a column depicting which session we can use a for loop:
dfs = []
for framename in data.keys():
temp_df = data[framename]
temp_df['Session'] = framename
dfs.append(temp_df)
df = pd.concat(dfs)Code language: Python (python)In the code above, we start by creating a list and continue by looping through the keys in the list of dataframes. Finally, we create a temporary dataframe and take the sheet name and add it in the column ‘Session’.
Pandas Read Excel: How to Read All Sheets
Now, it is, of course, possible that when we want to read multiple sheets we also want to read all the sheets in the Excel file. That is, if we want to use read_excel to load all sheets from an Excel file to a dataframe it is possible. When reading multiple sheets and we want all sheets we can set the parameter sheet_name to None.
all_sheets_df = pd.read_excel('example_sheets1.xlsx', sheet_name=None)Code language: Python (python)Pandas Read Excel: Reading Many Excel Files
In this section, of the Pandas read excel tutorial, we will learn how to load many files into a Pandas dataframe because, in some cases, we may have a lot of Excel files containing data from, let’s say, different experiments. In Python, we can use the modules os and fnmatch to read all files in a directory. Finally, we use list comprehension to use read_excel on all files we found:
import os, fnmatch
xlsx_files = fnmatch.filter(os.listdir('.'), '*concat*.xlsx')
dfs = [pd.read_excel(xlsx_file) for xlsx_file in xlsx_files]Code language: Python (python)If it makes sense we can, again, use the function concat to merge the dataframes:
df = pd.concat(dfs, sort=False)Code language: Python (python)There are other methods for reading many Excel files and merging them. We can, for instance, use the module glob together with Pandas concat to read multiple xlsx files:
import glob
list_of_xlsx = glob.glob('./*concat*.xlsx')
df = pd.concat(list_of_xlsx)Code language: Python (python)Note, the files in this example, where we read multiple xlsx files using Pandas, are located here. They are named example_concat.xlsx, example_concat1.xlsx, and example_concat3.xlsx and should be added to the same directory as the Python script. Another option, of course, is to add the file path to the files. E.g., if we want to read multiple Excel files, using Pandas read_excel method, and they are stored in a directory called “SimData” we would do as follows:
import glob
list_of_xlsx = glob.glob('./SimData/*concat*.xlsx')
df = pd.concat(list_of_xlsx)Code language: Python (python)Setting the Data Type for Data or Columns
If we need to, we can also, set the data type for the columns when reading Excel files using Pandas. Let’s use Pandas to read the example_sheets1.xlsx again. In the Pandas read_excel example below we use the dtype parameter to set the data type of some of the columns.
df = pd.read_excel('example_sheets1.xlsx',sheet_name='Session1',
header=1,dtype={'Names':str,'ID':str,
'Mean':int, 'Session':str})Code language: Python (python)We can use the method info to see the what data types the different columns have:
df.info()Code language: Python (python)
Writing Pandas Dataframes to Excel
Excel files can, of course, be created in Python using Pandas to_excel method. In this section of the post, we will learn how to create an excel file using Pandas. First, before writing an Excel file, we will create a dataframe containing some variables. Before that, we need to import Pandas:
import pandas as pdCode language: Python (python)The next step is to create the dataframe. We will create the dataframe using a dictionary. The keys will be the column names and the values will be lists containing our data:
df = pd.DataFrame({'Names':['Andreas', 'George', 'Steve',
'Sarah', 'Joanna', 'Hanna'],
'Age':[21, 22, 20, 19, 18, 23]})Code language: Python (python)In this Pandas write to Excel example, we will write the dataframe to an Excel file using the to_excel method. Noteworthy, when using Pandas to_excel in the code chunk below, we don’t use any parameters.
df.to_excel('NamesAndAges.xlsx')Code language: Python (python)In the Excel file created when using Pandas to_excel is shown below. Evidently, if we don’t use the parameter sheet_name we get the default sheet name, ‘Sheet1’. Now, we can also see that we get a new column in our Excel file containing numbers. These are the index from the dataframe.
If we want our sheet to be named something else and we don’t want the index column we can add the following argument and parameters when we use Pandas to write to Excel:
df.to_excel('NamesAndAges.xlsx', sheet_name='Names and Ages', index=False)Code language: Python (python)Writing Multiple Pandas Dataframes to an Excel File:
In this section, we are going to use Pandas ExcelWriter and Pandas to_excel to write multiple Pandas dataframes to one Excel file. That is if we happen to have many dataframes that we want to store in one Excel file but on different sheets, we can do this easily. However, we need to use Pandas ExcelWriter now:
df1 = pd.DataFrame({'Names': ['Andreas', 'George', 'Steve',
'Sarah', 'Joanna', 'Hanna'],
'Age':[21, 22, 20, 19, 18, 23]})
df2 = pd.DataFrame({'Names': ['Pete', 'Jordan', 'Gustaf',
'Sophie', 'Sally', 'Simone'],
'Age':[22, 21, 19, 19, 29, 21]})
df3 = pd.DataFrame({'Names': ['Ulrich', 'Donald', 'Jon',
'Jessica', 'Elisabeth', 'Diana'],
'Age':[21, 21, 20, 19, 19, 22]})
dfs = {'Group1':df1, 'Group2':df2, 'Group3':df3}
writer = pd.ExcelWriter('NamesAndAges.xlsx', engine='xlsxwriter')
for sheet_name in dfs.keys():
dfs[sheet_name].to_excel(writer, sheet_name=sheet_name, index=False)
writer.save()Code language: Python (python)In the code above, we create 3 dataframes and then we continue to put them in a dictionary. Note, the keys are the sheet names and the cell names are the dataframes. After this is done we create a writer object using the xlsxwriter engine. We then continue by looping through the keys (i.e., sheet names) and add each sheet. Finally, the file is saved. Note, the final step is important as leaving this out will not give you the intended results.
Of course, there are other ways to store data. One of them is using JSON files. See the latest tutorial on how to read and write JSON files using Pandas to learn about one way to load and save data in the JSON format.
More resources on how to load data in different formats:
- How to read and write CSV files using Pandas
- How to read and write SPSS files using Python
Summary: How to Work With Excel Files using Pandas
That was it! In this post, we have learned a lot! We have, among other things, learned how to:
- Read Excel files and Spreadsheets using read_excel
- Load Excel files to dataframes:
- Read Excel sheets and skip rows
- Merging many sheets to a dataframe
- Loading many Excel files into one dataframe
- Load Excel files to dataframes:
- Write a dataframe to an Excel file
- Taking many dataframes and writing them to one Excel file with many sheets
Leave a comment below if you have any requests or suggestions on what should be covered next! Check the post A Basic Pandas Dataframe Tutorial for Beginners to learn more about working with Pandas dataframe. That is after you have loaded them from a file (e.g., Excel spreadsheets)